Identification of regulators of chaperone-mediated autophagy
- PMID: 20797626
- PMCID: PMC2945256
- DOI: 10.1016/j.molcel.2010.08.004
Identification of regulators of chaperone-mediated autophagy
Abstract
Chaperone-mediated autophagy (CMA) is a selective mechanism for the degradation of cytosolic proteins in lysosomes that contributes to cellular quality control and becomes an additional source of amino acids when nutrients are scarce. A chaperone complex delivers CMA substrates to a receptor protein at the lysosomal membrane that assembles into multimeric translocation complexes. However, the mechanisms regulating this process remain, for the most part, unknown. In this work, we have identified two regulatory proteins, GFAP and EF1alpha, that mediate a previously unknown inhibitory effect of GTP on CMA. GFAP stabilizes the multimeric translocation complex against chaperone-mediated disassembly, whereas GTP-mediated release of EF1alpha from the lysosomal membrane promotes self-association of GFAP, disassembly of the CMA translocation complex, and the consequent decrease in CMA. The dynamic interactions of these two proteins at the lysosomal membrane unveil now a role for GTP as a negative regulator of CMA.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
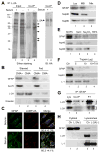
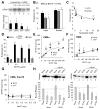

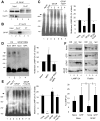
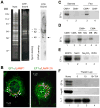

Comment in
-
GTP: gatekeeper for autophagy.Mol Cell. 2010 Aug 27;39(4):485-6. doi: 10.1016/j.molcel.2010.08.005. Mol Cell. 2010. PMID: 20797620
Similar articles
-
Entering the lysosome through a transient gate by chaperone-mediated autophagy.Autophagy. 2008 Nov;4(8):1101-3. doi: 10.4161/auto.7150. Epub 2008 Nov 8. Autophagy. 2008. PMID: 18927485 Free PMC article.
-
Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy.EMBO J. 2006 Sep 6;25(17):3921-33. doi: 10.1038/sj.emboj.7601283. Epub 2006 Aug 17. EMBO J. 2006. PMID: 16917501 Free PMC article.
-
Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy.Mol Cell. 2015 Jul 16;59(2):270-84. doi: 10.1016/j.molcel.2015.05.030. Epub 2015 Jun 25. Mol Cell. 2015. PMID: 26118642 Free PMC article.
-
Chaperone-mediated autophagy: roles in disease and aging.Cell Res. 2014 Jan;24(1):92-104. doi: 10.1038/cr.2013.153. Epub 2013 Nov 26. Cell Res. 2014. PMID: 24281265 Free PMC article. Review.
-
Chaperone-mediated autophagy.Autophagy. 2007 Jul-Aug;3(4):295-9. doi: 10.4161/auto.4144. Epub 2007 Jul 15. Autophagy. 2007. PMID: 17404494 Review.
Cited by
-
Chaperone-mediated autophagy: roles in neuroprotection.Neurosci Bull. 2015 Aug;31(4):452-8. doi: 10.1007/s12264-015-1540-x. Epub 2015 Jul 23. Neurosci Bull. 2015. PMID: 26206599 Free PMC article. Review.
-
Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice.Nat Commun. 2021 Apr 14;12(1):2238. doi: 10.1038/s41467-021-22501-9. Nat Commun. 2021. PMID: 33854069 Free PMC article.
-
Astrocyte-Neuron Interactions in Alzheimer's Disease.Adv Neurobiol. 2024;39:345-382. doi: 10.1007/978-3-031-64839-7_14. Adv Neurobiol. 2024. PMID: 39190082 Review.
-
PRRSV GP5 inhibits the antivirus effects of chaperone-mediated autophagy by targeting LAMP2A.mBio. 2024 Aug 14;15(8):e0053224. doi: 10.1128/mbio.00532-24. Epub 2024 Jun 28. mBio. 2024. PMID: 38940560 Free PMC article.
-
The Role of Chaperone-Mediated Autophagy in Bortezomib Resistant Multiple Myeloma.Cells. 2021 Dec 8;10(12):3464. doi: 10.3390/cells10123464. Cells. 2021. PMID: 34943972 Free PMC article.
References
-
- Boran MS, Garcia A. The cyclic GMP-protein kinase G pathway regulates cytoskeleton dynamics and motility in astrocytes. J Neurochem. 2007;102:216–230. - PubMed
-
- Coppard NJ, Clark BF, Cramer F. Methylation of elongation factor 1 alpha in mouse 3T3B and 3T3B/SV40 cells. FEBS Lett. 1983;164:330–334. - PubMed
-
- Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. - PubMed
-
- Cuervo A, Dice J. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113:4441–4450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

