A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes
- PMID: 20797779
- PMCID: PMC2954371
- DOI: 10.1016/j.cell.2010.07.038
A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes
Abstract
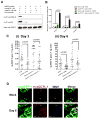
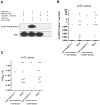
West Nile virus (WNV) is the most common arthropod-borne flavivirus in the United States; however, the vector ligand(s) that participate in infection are not known. We now show that an Aedes aegypti C-type lectin, mosGCTL-1, is induced by WNV, interacts with WNV in a calcium-dependent manner, and facilitates infection in vivo and in vitro. A mosquito homolog of human CD45 in A. aegypti, designated mosPTP-1, recruits mosGCTL-1 to enable viral attachment to cells and to enhance viral entry. In vivo experiments show that mosGCTL-1 and mosPTP-1 function as part of the same pathway and are critical for WNV infection of mosquitoes. A similar phenomenon was also observed in Culex quinquefasciatus, a natural vector of WNV, further demonstrating that these genes participate in WNV infection. During the mosquito blood-feeding process, WNV infection was blocked in vivo with mosGCTL-1 antibodies. A molecular understanding of flaviviral-arthropod interactions may lead to strategies to control viral dissemination in nature.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
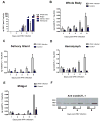
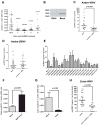
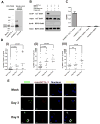
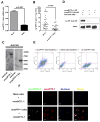
References
-
- Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. - PubMed
-
- Arnold JN, Dwek RA, Rudd PM, Sim RB. Mannan binding lectin and its interaction with immunoglobulins in health and in disease. Immunol Lett. 2006;106:103–110. - PubMed
-
- Baldwin TA, Ostergaard HL. Developmentally regulated changes in glucosidase II association with, and carbohydrate content of, the protein tyrosine phosphatase CD45. J Immunol. 2001;167:3829–3835. - PubMed
-
- Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

