The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation
- PMID: 20797886
- PMCID: PMC4158944
- DOI: 10.1016/j.molcel.2010.08.011
The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation
Abstract
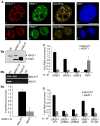
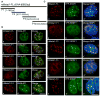
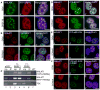
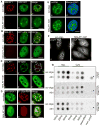
Alternative splicing (AS) of pre-mRNA is utilized by higher eukaryotes to achieve increased transcriptome and proteomic complexity. The serine/arginine (SR) splicing factors regulate tissue- or cell-type-specific AS in a concentration- and phosphorylation-dependent manner. However, the mechanisms that modulate the cellular levels of active SR proteins remain to be elucidated. In the present study, we provide evidence for a role for the long nuclear-retained regulatory RNA (nrRNA), MALAT1 in AS regulation. MALAT1 interacts with SR proteins and influences the distribution of these and other splicing factors in nuclear speckle domains. Depletion of MALAT1 or overexpression of an SR protein changes the AS of a similar set of endogenous pre-mRNAs. Furthermore, MALAT1 regulates cellular levels of phosphorylated forms of SR proteins. Taken together, our results suggest that MALAT1 regulates AS by modulating the levels of active SR proteins. Our results further highlight the role for an nrRNA in the regulation of gene expression.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

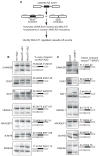
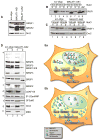
Comment in
-
Long noncoding RNAs add another layer to pre-mRNA splicing regulation.Mol Cell. 2010 Sep 24;39(6):833-4. doi: 10.1016/j.molcel.2010.09.003. Mol Cell. 2010. PMID: 20864030
Similar articles
-
SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles.Mol Biol Cell. 2012 Sep;23(18):3694-706. doi: 10.1091/mbc.E12-03-0206. Epub 2012 Aug 1. Mol Biol Cell. 2012. PMID: 22855529 Free PMC article.
-
Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function.Biochem J. 2015 Mar 1;466(2):311-22. doi: 10.1042/BJ20141373. Biochem J. 2015. PMID: 25529026 Free PMC article.
-
Nuclear ALG-2 protein interacts with Ca2+ homeostasis endoplasmic reticulum protein (CHERP) Ca2+-dependently and participates in regulation of alternative splicing of inositol trisphosphate receptor type 1 (IP3R1) pre-mRNA.J Biol Chem. 2013 Nov 15;288(46):33361-75. doi: 10.1074/jbc.M113.497479. Epub 2013 Sep 27. J Biol Chem. 2013. PMID: 24078636 Free PMC article.
-
Plant serine/arginine-rich proteins: versatile players in RNA processing.Planta. 2023 May 5;257(6):109. doi: 10.1007/s00425-023-04132-0. Planta. 2023. PMID: 37145304 Review.
-
Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):875-89. doi: 10.1002/wrna.98. Epub 2011 Jul 15. Wiley Interdiscip Rev RNA. 2011. PMID: 21766458 Review.
Cited by
-
Regulation of mammalian cell differentiation by long non-coding RNAs.EMBO Rep. 2012 Nov 6;13(11):971-83. doi: 10.1038/embor.2012.145. Epub 2012 Oct 16. EMBO Rep. 2012. PMID: 23070366 Free PMC article. Review.
-
Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients.Oncol Lett. 2015 Feb;9(2):955-963. doi: 10.3892/ol.2014.2730. Epub 2014 Nov 24. Oncol Lett. 2015. PMID: 25624916 Free PMC article.
-
LncRNA TP53TG1 plays an anti-oncogenic role in cervical cancer by synthetically regulating transcriptome profile in HeLa cells.Front Genet. 2022 Oct 4;13:981030. doi: 10.3389/fgene.2022.981030. eCollection 2022. Front Genet. 2022. PMID: 36267418 Free PMC article.
-
Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles.RNA. 2012 Apr;18(4):738-51. doi: 10.1261/rna.028639.111. Epub 2012 Feb 21. RNA. 2012. PMID: 22355166 Free PMC article.
-
DeepLocRNA: an interpretable deep learning model for predicting RNA subcellular localization with domain-specific transfer-learning.Bioinformatics. 2024 Feb 1;40(2):btae065. doi: 10.1093/bioinformatics/btae065. Bioinformatics. 2024. PMID: 38317052 Free PMC article.
References
-
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. - PubMed
-
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. - PubMed
-
- Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol. 2004;78:37–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

