Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors
- PMID: 20798047
- PMCID: PMC2936598
- DOI: 10.1073/pnas.1006901107
Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors
Erratum in
- Proc Natl Acad Sci U S A. 2010 Nov 2;107(44):19132
Retraction in
-
Retraction for Lo et al., Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors.Proc Natl Acad Sci U S A. 2012 Jan 3;109(1):346. doi: 10.1073/pnas.1119641109. Epub 2011 Dec 27. Proc Natl Acad Sci U S A. 2012. PMID: 22203980 Free PMC article. No abstract available.
Abstract
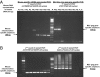
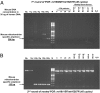
Chronic fatigue syndrome (CFS) is a serious systemic illness of unknown cause. A recent study identified DNA from a xenotropic murine leukemia virus-related virus (XMRV) in peripheral blood mononuclear cells (PBMCs) from 68 of 101 patients (67%) by nested PCR, as compared with 8 of 218 (3.7%) healthy controls. However, four subsequent reports failed to detect any murine leukemia virus (MLV)-related virus gene sequences in blood of CFS patients. We examined 41 PBMC-derived DNA samples from 37 patients meeting accepted diagnostic criteria for CFS and found MLV-like virus gag gene sequences in 32 of 37 (86.5%) compared with only 3 of 44 (6.8%) healthy volunteer blood donors. No evidence of mouse DNA contamination was detected in the PCR assay system or the clinical samples. Seven of 8 gag-positive patients tested again positive in a sample obtained nearly 15 y later. In contrast to the reported findings of near-genetic identity of all XMRVs, we identified a genetically diverse group of MLV-related viruses. The gag and env sequences from CFS patients were more closely related to those of polytropic mouse endogenous retroviruses than to those of XMRVs and were even less closely related to those of ecotropic MLVs. Further studies are needed to determine whether the same strong association with MLV-related viruses is found in other groups of patients with CFS, whether these viruses play a causative role in the development of CFS, and whether they represent a threat to the blood supply.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
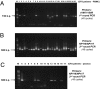
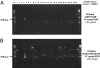
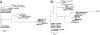
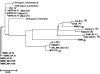
Comment in
-
Mouse retroviruses and chronic fatigue syndrome: Does X (or P) mark the spot?Proc Natl Acad Sci U S A. 2010 Sep 7;107(36):15666-7. doi: 10.1073/pnas.1007944107. Epub 2010 Aug 23. Proc Natl Acad Sci U S A. 2010. PMID: 20798036 Free PMC article. No abstract available.
-
Patients, patience, and the publication process.Proc Natl Acad Sci U S A. 2010 Sep 7;107(36):15661. doi: 10.1073/pnas.1012027107. Epub 2010 Aug 23. Proc Natl Acad Sci U S A. 2010. PMID: 20798042 Free PMC article. No abstract available.
-
Chronic fatigue syndrome: xenotropic murine leukemia virus-related virus, murine leukemia virus, both, or neither?Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):E161; author reply E163-4. doi: 10.1073/pnas.1012441107. Epub 2010 Sep 30. Proc Natl Acad Sci U S A. 2010. PMID: 20884850 Free PMC article. No abstract available.
-
Possible widespread low-level occurrence of murine leukemia virus-related gene sequences in humans.Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):E162; author reply E163-4. doi: 10.1073/pnas.1012930107. Epub 2010 Sep 30. Proc Natl Acad Sci U S A. 2010. PMID: 20884851 Free PMC article. No abstract available.
References
-
- Fukuda K, et al. International Chronic Fatigue Syndrome Study Group The chronic fatigue syndrome: A comprehensive approach to its definition and study. Ann Intern Med. 1994;121:953–959. - PubMed
-
- Komaroff AL. Is human herpesvirus-6 a trigger for chronic fatigue syndrome? J Clin Virol. 2006;37(Suppl 1):S39–S46. - PubMed
-
- Lombardi VC, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–589. - PubMed
-
- Fischer N, et al. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J Clin Virol. 2008;43:277–283. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

