Nanopore DNA sequencing with MspA
- PMID: 20798343
- PMCID: PMC2941267
- DOI: 10.1073/pnas.1001831107
Nanopore DNA sequencing with MspA
Abstract
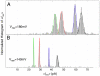
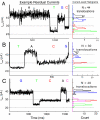
Nanopore sequencing has the potential to become a direct, fast, and inexpensive DNA sequencing technology. The simplest form of nanopore DNA sequencing utilizes the hypothesis that individual nucleotides of single-stranded DNA passing through a nanopore will uniquely modulate an ionic current flowing through the pore, allowing the record of the current to yield the DNA sequence. We demonstrate that the ionic current through the engineered Mycobacterium smegmatis porin A, MspA, has the ability to distinguish all four DNA nucleotides and resolve single-nucleotides in single-stranded DNA when double-stranded DNA temporarily holds the nucleotides in the pore constriction. Passing DNA with a series of double-stranded sections through MspA provides proof of principle of a simple DNA sequencing method using a nanopore. These findings highlight the importance of MspA in the future of nanopore sequencing.
Conflict of interest statement
Conflict of interest statement: The authors declare a conflict of interest (such as defined by PNAS policy). J.H.G., M.N., T.Z.B., and M.P. have filed a patent on the use of MspA for single-molecule analysis. I.M.D., M.D.C, and J.H.G. have filed a provisional patent on the sequencing strategy presented herein.
Figures



References
-
- Goldstein DB. Common Genetic Variation and Human Traits. N Engl J Med. 2009;360(17):1696–1698. - PubMed
-
- Hirschhorn JN. Genomewide association studies—Illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. - PubMed
-
- Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462(7275):886–890. - PubMed
-
- Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. In: Ausubel Frederick M, et al., editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 2008. Chapter 7. - PubMed
-
- Fuller CW, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27(11):1013–1023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

