Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population
- PMID: 20798349
- PMCID: PMC2941333
- DOI: 10.1073/pnas.1004381107
Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population
Abstract
The Ashkenazi Jewish (AJ) population has long been viewed as a genetic isolate, yet it is still unclear how population bottlenecks, admixture, or positive selection contribute to its genetic structure. Here we analyzed a large AJ cohort and found higher linkage disequilibrium (LD) and identity-by-descent relative to Europeans, as expected for an isolate. However, paradoxically we also found higher genetic diversity, a sign of an older or more admixed population but not of a long-term isolate. Recent reports have reaffirmed that the AJ population has a common Middle Eastern origin with other Jewish Diaspora populations, but also suggest that the AJ population, compared with other Jews, has had the most European admixture. Our analysis indeed revealed higher European admixture than predicted from previous Y-chromosome analyses. Moreover, we also show that admixture directly correlates with high LD, suggesting that admixture has increased both genetic diversity and LD in the AJ population. Additionally, we applied extended haplotype tests to determine whether positive selection can account for the level of AJ-prevalent diseases. We identified genomic regions under selection that account for lactose and alcohol tolerance, and although we found evidence for positive selection at some AJ-prevalent disease loci, the higher incidence of the majority of these diseases is likely the result of genetic drift following a bottleneck. Thus, the AJ population shows evidence of past founding events; however, admixture and selection have also strongly influenced its current genetic makeup.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
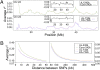
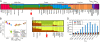


References
-
- Ostrer H. A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2:891–898. - PubMed
-
- Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61:233–247. - PubMed
-
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

