Stimulation of DNA Glycosylase Activities by XPC Protein Complex: Roles of Protein-Protein Interactions
- PMID: 20798892
- PMCID: PMC2925305
- DOI: 10.4061/2010/805698
Stimulation of DNA Glycosylase Activities by XPC Protein Complex: Roles of Protein-Protein Interactions
Abstract
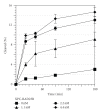
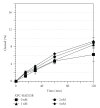
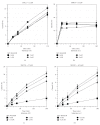
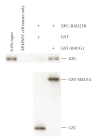
We showed that XPC complex, which is a DNA damage detector for nucleotide excision repair, stimulates activity of thymine DNA glycosylase (TDG) that initiates base excision repair. XPC appeared to facilitate the enzymatic turnover of TDG by promoting displacement from its own product abasic site, although the precise mechanism underlying this stimulation has not been clarified. Here we show that XPC has only marginal effects on the activity of E. coli TDG homolog (EcMUG), which remains bound to the abasic site like human TDG but does not significantly interacts with XPC. On the contrary, XPC significantly stimulates the activities of sumoylated TDG and SMUG1, both of which exhibit quite different enzymatic kinetics from unmodified TDG but interact with XPC. These results point to importance of physical interactions for stimulation of DNA glycosylases by XPC and have implications in the molecular mechanisms underlying mutagenesis and carcinogenesis in XP-C patients.
Figures



Similar articles
-
Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase.EMBO J. 2003 Jan 2;22(1):164-73. doi: 10.1093/emboj/cdg016. EMBO J. 2003. PMID: 12505994 Free PMC article.
-
Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation.Curr Biol. 2005 Apr 12;15(7):616-23. doi: 10.1016/j.cub.2005.02.054. Curr Biol. 2005. PMID: 15823533
-
Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells.Genes Dev. 2017 Apr 15;31(8):830-844. doi: 10.1101/gad.295741.116. Genes Dev. 2017. PMID: 28512237 Free PMC article.
-
Structural and mutation studies of two DNA demethylation related glycosylases: MBD4 and TDG.Biophysics (Nagoya-shi). 2014 Oct 18;10:63-8. doi: 10.2142/biophysics.10.63. eCollection 2014. Biophysics (Nagoya-shi). 2014. PMID: 27493500 Free PMC article. Review.
-
[Uracil-DNA glycosylases].Postepy Biochem. 2008;54(4):362-70. Postepy Biochem. 2008. PMID: 19248582 Review. Polish.
Cited by
-
Global-genome Nucleotide Excision Repair Controlled by Ubiquitin/Sumo Modifiers.Front Genet. 2016 Apr 28;7:68. doi: 10.3389/fgene.2016.00068. eCollection 2016. Front Genet. 2016. PMID: 27200078 Free PMC article. Review.
-
DNA Oxidation and Excision Repair Pathways.Int J Mol Sci. 2019 Dec 3;20(23):6092. doi: 10.3390/ijms20236092. Int J Mol Sci. 2019. PMID: 31816862 Free PMC article. Review.
-
Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.DNA Repair (Amst). 2021 Nov;107:103194. doi: 10.1016/j.dnarep.2021.103194. Epub 2021 Jul 29. DNA Repair (Amst). 2021. PMID: 34428697 Free PMC article.
-
PPRC1, but not PGC-1α, levels directly correlate with expression of mitochondrial proteins in human dermal fibroblasts.Genet Mol Biol. 2020 Jul 3;43(1 suppl. 1):e20190083. doi: 10.1590/1678-4685-GMB-2019-0083. Genet Mol Biol. 2020. PMID: 32639509 Free PMC article.
-
The Multiple Cellular Roles of SMUG1 in Genome Maintenance and Cancer.Int J Mol Sci. 2021 Feb 17;22(4):1981. doi: 10.3390/ijms22041981. Int J Mol Sci. 2021. PMID: 33671338 Free PMC article. Review.
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edition. Washington, DC, USA: ASM Press; 2006.
-
- McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annual Review of Biochemistry. 1999;68:255–285. - PubMed
-
- Schärer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. BioEssays. 2001;23(3):270–281. - PubMed
-
- Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JHJ. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. New York, NY, USA: McGraw-Hill; 2001. pp. 677–703.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

