Structure characterization of the 26S proteasome
- PMID: 20800708
- PMCID: PMC3010250
- DOI: 10.1016/j.bbagrm.2010.08.008
Structure characterization of the 26S proteasome
Abstract
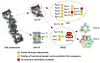
In all eukaryotic cells, 26S proteasome plays an essential role in the process of ATP-dependent protein degradation. In this review, we focus on structure characterization of the 26S proteasome. Although the progress towards a high-resolution structure of the 26S proteasome has been slow, the recently solved structures of various proteasomal subcomplexes have greatly enhanced our understanding of this large machinery. In addition to having an ATP-dependent proteolytic function, the 26S proteasome is also involved in many non-proteolytic cellular activities, which are often mediated by subunits in its 19S regulatory complex. Thus, we include a detailed discussion of the structures of 19S subunits, including proteasomal ATPases, ubiquitin receptors, deubiquitinating enzymes and subunits that contain PCI domain. This article is part of a Special Issue entitled The 26S Proteasome: When degradation is just not enough!
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures

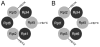
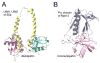


References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
-
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

