Vertical distribution of ammonia-oxidizing crenarchaeota and methanogens in the epipelagic waters of Lake Kivu (Rwanda-Democratic Republic of the Congo)
- PMID: 20802065
- PMCID: PMC2953034
- DOI: 10.1128/AEM.02864-09
Vertical distribution of ammonia-oxidizing crenarchaeota and methanogens in the epipelagic waters of Lake Kivu (Rwanda-Democratic Republic of the Congo)
Abstract
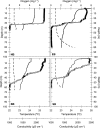
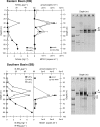
Four stratified basins in Lake Kivu (Rwanda-Democratic Republic of the Congo) were sampled in March 2007 to investigate the abundance, distribution, and potential biogeochemical role of planktonic archaea. We used fluorescence in situ hybridization with catalyzed-reported deposition microscopic counts (CARD-FISH), denaturing gradient gel electrophoresis (DGGE) fingerprinting, and quantitative PCR (qPCR) of signature genes for ammonia-oxidizing archaea (16S rRNA for marine Crenarchaeota group 1.1a [MCG1] and ammonia monooxygenase subunit A [amoA]). Abundance of archaea ranged from 1 to 4.5% of total DAPI (4',6-diamidino-2-phenylindole) counts with maximal concentrations at the oxic-anoxic transition zone (∼50-m depth). Phylogenetic analysis of the archaeal planktonic community revealed a higher level of richness of crenarchaeal 16S rRNA gene sequences (21 of the 28 operational taxonomic units [OTUs] identified [75%]) over euryarchaeotal ones (7 OTUs). Sequences affiliated with the kingdom Euryarchaeota were mainly recovered from the anoxic water compartment and mostly grouped into methanogenic lineages (Methanosarcinales and Methanocellales). In turn, crenarchaeal phylotypes were recovered throughout the sampled epipelagic waters (0- to 100-m depth), with clear phylogenetic segregation along the transition from oxic to anoxic water masses. Thus, whereas in the anoxic hypolimnion crenarchaeotal OTUs were mainly assigned to the miscellaneous crenarchaeotic group, the OTUs from the oxic-anoxic transition and above belonged to Crenarchaeota groups 1.1a and 1.1b, two lineages containing most of the ammonia-oxidizing representatives known so far. The concomitant vertical distribution of both nitrite and nitrate maxima and the copy numbers of both MCG1 16S rRNA and amoA genes suggest the potential implication of Crenarchaeota in nitrification processes occurring in the epilimnetic waters of the lake.
Figures



References
-
- American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed. APHA, Washington, DC.
-
- Auguet, J. C., A. Barberan, and E. O. Casamayor. 2010. Global ecological patterns in uncultured Archaea. ISME J. 4:182-190. - PubMed
-
- Auguet, J. C., and E. O. Casamayor. 2008. A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ. Microbiol. 10:1080-1086. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

