Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization
- PMID: 20802066
- PMCID: PMC2976205
- DOI: 10.1128/AEM.00675-10
Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization
Abstract
Human milk oligosaccharides (HMOs) are the third-largest solid component of milk. Their structural complexity renders them nondigestible to the host but liable to hydrolytic enzymes of the infant colonic microbiota. Bifidobacteria and, frequently, Bifidobacterium longum strains predominate the colonic microbiota of exclusively breast-fed infants. Among the three recognized subspecies of B. longum, B. longum subsp. infantis achieves high levels of cell growth on HMOs and is associated with early colonization of the infant gut. The B. longum subsp. infantis ATCC 15697 genome features five distinct gene clusters with the predicted capacity to bind, cleave, and import milk oligosaccharides. Comparative genomic hybridizations (CGHs) were used to associate genotypic biomarkers among 15 B. longum strains exhibiting various HMO utilization phenotypes and host associations. Multilocus sequence typing provided taxonomic subspecies designations and grouped the strains between B. longum subsp. infantis and B. longum subsp. longum. CGH analysis determined that HMO utilization gene regions are exclusively conserved across all B. longum subsp. infantis strains capable of growth on HMOs and have diverged in B. longum subsp. longum strains that cannot grow on HMOs. These regions contain fucosidases, sialidases, glycosyl hydrolases, ABC transporters, and family 1 solute binding proteins and are likely needed for efficient metabolism of HMOs. Urea metabolism genes and their activity were exclusively conserved in B. longum subsp. infantis. These results imply that the B. longum has at least two distinct subspecies: B. longum subsp. infantis, specialized to utilize milk carbon, and B. longum subsp. longum, specialized for plant-derived carbon metabolism.
Figures
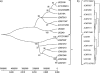
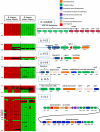
References
-
- Ahrens, K. 2009. Consumption of human milk oligosaccharides by Bifidobacterium. MS thesis. University of California—Davis, Davis, CA.
-
- Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. - PubMed
-
- Cai, H., B. T. Rodriguez, W. Zhang, J. R. Broadbent, and J. L. Steele. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153:2655-2665. - PubMed
-
- Chaturvedi, P., C. D. Warren, C. R. Buescher, L. K. Pickering, and D. S. Newburg. 2001. Survival of human milk oligosaccharides in the intestine of infants, p. 315-323. In D. S. Newburg (ed.), Bioactive components of human milk. Springer, New York, NY. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

