Proteolytic enzyme production by strains of the insect pathogen xenorhabdus and characterization of an early-log-phase-secreted protease as a potential virulence factor
- PMID: 20802071
- PMCID: PMC2953030
- DOI: 10.1128/AEM.01567-10
Proteolytic enzyme production by strains of the insect pathogen xenorhabdus and characterization of an early-log-phase-secreted protease as a potential virulence factor
Abstract
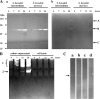
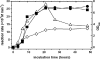
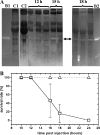
As a comparison to a similar study on Photorhabdus strains, 15 Xenorhabdus bacterial strains and secondary phenotypic variants of two strains were screened for proteolytic activity by five detection methods. Although the number and intensity of proteolytic activities were different, every strain was positive for proteolytic activity by several tests. Zymography following native PAGE detected two groups of activities with different substrate affinities and a higher and lower electrophoretic mobility that were distinguished as activity 1 and 2, respectively. Zymography following SDS-PAGE resolved three activities, which were provisionally named proteases A, B, and C. Only protease B, an ∼55-kDa enzyme, was produced by every strain. This enzyme exhibited higher affinity to the gelatin substrate than to the casein substrate. Of the chromogenic substrates used, three were hydrolyzed: furylacryloyl-Ala-Leu-Val-Tyr (Fua-ALVY), Fua-LGPA (LGPA is Leu-Gly-Pro-Ala) (a substrate for collagen peptidases), and succinyl-Ala-Ala-Pro-Phe-thiobenzyl (Succ-AAPF-SBzl). All but the Fua-LGPA-ase activity seemed to be from secreted enzymes. According to their substrate preference profiles and inhibitor sensitivities, at least six such proteolytic enzymes could be distinguished in the culture medium of Xenorhabdus strains. The proteolytic enzyme that was secreted the earliest, protease B and the Succ-AAPF-SBzl-hydrolyzing enzyme, appeared from the early logarithmic phase of growth. Protease B could also be detected in the hemolymph of Xenorhabdus-infected Galleria mellonella larvae from 15 h postinfection. The purified protease B hydrolyzed in vitro seven proteins in the hemolymph of Manduca sexta that were also cleaved by PrtA peptidase from Photorhabdus. The N-terminal sequence of protease B showed similarity to a 55-kDa serralysin type metalloprotease in Xenorhabdus nematophila, which had been identified as an orthologue of Photorhabdus PrtA peptidase.
Figures


Similar articles
-
Comparison of proteolytic activities produced by entomopathogenic Photorhabdus bacteria: strain- and phase-dependent heterogeneity in composition and activity of four enzymes.Appl Environ Microbiol. 2004 Dec;70(12):7311-20. doi: 10.1128/AEM.70.12.7311-7320.2004. Appl Environ Microbiol. 2004. PMID: 15574931 Free PMC article.
-
Enzymic characterization with progress curve analysis of a collagen peptidase from an enthomopathogenic bacterium, Photorhabdus luminescens.Biochem J. 2004 May 1;379(Pt 3):633-40. doi: 10.1042/BJ20031116. Biochem J. 2004. PMID: 14744262 Free PMC article.
-
Examination of Xenorhabdus nematophila lipases in pathogenic and mutualistic host interactions reveals a role for xlpA in nematode progeny production.Appl Environ Microbiol. 2010 Jan;76(1):221-9. doi: 10.1128/AEM.01715-09. Epub 2009 Oct 30. Appl Environ Microbiol. 2010. PMID: 19880652 Free PMC article.
-
Flagellar Regulation and Virulence in the Entomopathogenic Bacteria-Xenorhabdus nematophila and Photorhabdus luminescens.Curr Top Microbiol Immunol. 2017;402:39-51. doi: 10.1007/82_2016_53. Curr Top Microbiol Immunol. 2017. PMID: 28091933 Review.
-
The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: a review.Parasit Vectors. 2020 Jul 29;13(1):376. doi: 10.1186/s13071-020-04236-6. Parasit Vectors. 2020. PMID: 32727530 Free PMC article. Review.
Cited by
-
Purification and properties of an insecticidal metalloprotease produced by Photorhabdus luminescens strain 0805-P5G, the entomopathogenic nematode symbiont.Int J Mol Sci. 2012 Dec 21;14(1):308-21. doi: 10.3390/ijms14010308. Int J Mol Sci. 2012. PMID: 23344035 Free PMC article.
-
Comparison of Xenorhabdus bovienii bacterial strain genomes reveals diversity in symbiotic functions.BMC Genomics. 2015 Nov 2;16:889. doi: 10.1186/s12864-015-2000-8. BMC Genomics. 2015. PMID: 26525894 Free PMC article.
-
Understanding pine wilt disease: roles of the pine endophytic bacteria and of the bacteria carried by the disease-causing pinewood nematode.Microbiologyopen. 2017 Apr;6(2):e00415. doi: 10.1002/mbo3.415. Epub 2016 Oct 26. Microbiologyopen. 2017. PMID: 27785885 Free PMC article. Review.
-
Efficient Keratinolysis of Poultry Feather Waste by the Halotolerant Keratinase from Salicola Marasensis.Iran J Pharm Res. 2019 Fall;18(4):1862-1870. doi: 10.22037/ijpr.2019.111710.13312. Iran J Pharm Res. 2019. PMID: 32184853 Free PMC article.
-
FliZ is a global regulatory protein affecting the expression of flagellar and virulence genes in individual Xenorhabdus nematophila bacterial cells.PLoS Genet. 2013 Oct;9(10):e1003915. doi: 10.1371/journal.pgen.1003915. Epub 2013 Oct 31. PLoS Genet. 2013. PMID: 24204316 Free PMC article.
References
-
- Akhurst, R. J. 1980. Morphological and functional dimorphism in Xenorhabdus spp., bacteria symbiotically associated with the insect parasitic nematodes. J. Invertebr. Pathol. 62:68-72.
-
- Boemare, N. E., and R. J. Akhurst. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus ssp. (Enterobacteriaceae). J. Gen. Microbiol. 134:751-761. - PubMed
-
- Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255.
-
- Boemare, N. 2002. Biology, taxonomy and systematics of Photorhabdus and Xenorhabdus, p. 35-56. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, Oxon, United Kingdom.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

