Reference-unbiased copy number variant analysis using CGH microarrays
- PMID: 20802225
- PMCID: PMC2978381
- DOI: 10.1093/nar/gkq730
Reference-unbiased copy number variant analysis using CGH microarrays
Abstract
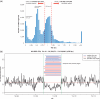
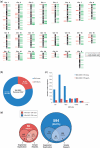
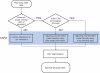
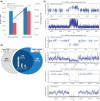
Comparative genomic hybridization (CGH) microarrays have been used to determine copy number variations (CNVs) and their effects on complex diseases. Detection of absolute CNVs independent of genomic variants of an arbitrary reference sample has been a critical issue in CGH array experiments. Whole genome analysis using massively parallel sequencing with multiple ultra-high resolution CGH arrays provides an opportunity to catalog highly accurate genomic variants of the reference DNA (NA10851). Using information on variants, we developed a new method, the CGH array reference-free algorithm (CARA), which can determine reference-unbiased absolute CNVs from any CGH array platform. The algorithm enables the removal and rescue of false positive and false negative CNVs, respectively, which appear due to the effects of genomic variants of the reference sample in raw CGH array experiments. We found that the CARA remarkably enhanced the accuracy of CGH array in determining absolute CNVs. Our method thus provides a new approach to interpret CGH array data for personalized medicine.
Figures

Similar articles
-
The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome.BMC Med Genomics. 2011 Jan 27;4:16. doi: 10.1186/1755-8794-4-16. BMC Med Genomics. 2011. PMID: 21272361 Free PMC article.
-
Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing.Nat Genet. 2010 May;42(5):400-5. doi: 10.1038/ng.555. Epub 2010 Apr 4. Nat Genet. 2010. PMID: 20364138 Free PMC article.
-
Evaluation of copy number variation detection for a SNP array platform.BMC Bioinformatics. 2014 Feb 21;15:50. doi: 10.1186/1471-2105-15-50. BMC Bioinformatics. 2014. PMID: 24555668 Free PMC article.
-
Novel applications of array comparative genomic hybridization in molecular diagnostics.Expert Rev Mol Diagn. 2018 Jun;18(6):531-542. doi: 10.1080/14737159.2018.1479253. Epub 2018 May 31. Expert Rev Mol Diagn. 2018. PMID: 29848116 Review.
-
Copy number variations and stroke.Neurol Sci. 2016 Dec;37(12):1895-1904. doi: 10.1007/s10072-016-2658-y. Epub 2016 Jul 8. Neurol Sci. 2016. PMID: 27393281 Free PMC article. Review.
Cited by
-
Increased genome instability in human DNA segments with self-chains: homology-induced structural variations via replicative mechanisms.Hum Mol Genet. 2013 Jul 1;22(13):2642-51. doi: 10.1093/hmg/ddt113. Epub 2013 Mar 7. Hum Mol Genet. 2013. PMID: 23474816 Free PMC article.
-
The first Irish genome and ways of improving sequence accuracy.Genome Biol. 2010;11(9):132. doi: 10.1186/gb-2010-11-9-132. Epub 2010 Sep 7. Genome Biol. 2010. PMID: 20815917 Free PMC article.
-
tRNA gene copy number variation in humans.Gene. 2014 Feb 25;536(2):376-84. doi: 10.1016/j.gene.2013.11.049. Epub 2013 Dec 14. Gene. 2014. PMID: 24342656 Free PMC article.
-
Genetic association studies of copy-number variation: should assignment of copy number states precede testing?PLoS One. 2012;7(4):e34262. doi: 10.1371/journal.pone.0034262. Epub 2012 Apr 6. PLoS One. 2012. PMID: 22493684 Free PMC article.
-
Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals.Nat Genet. 2011 Jul 3;43(8):745-52. doi: 10.1038/ng.872. Nat Genet. 2011. PMID: 21725310
References
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

