Evolutionary history of chimpanzees inferred from complete mitochondrial genomes
- PMID: 20802239
- PMCID: PMC3108604
- DOI: 10.1093/molbev/msq227
Evolutionary history of chimpanzees inferred from complete mitochondrial genomes
Abstract
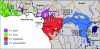
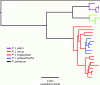
Investigations into the evolutionary history of the common chimpanzee, Pan troglodytes, have produced inconsistent results due to differences in the types of molecular data considered, the model assumptions employed, and the quantity and geographical range of samples used. We amplified and sequenced 24 complete P. troglodytes mitochondrial genomes from fecal samples collected at multiple study sites throughout sub-Saharan Africa. Using a "relaxed molecular clock," fossil calibrations, and 12 additional complete primate mitochondrial genomes, we analyzed the pattern and timing of primate diversification in a Bayesian framework. Our results support the recognition of four chimpanzee subspecies. Within P. troglodytes, we report a mean (95% highest posterior density [HPD]) time since most recent common ancestor (tMRCA) of 1.026 (0.811-1.263) Ma for the four proposed subspecies, with two major lineages. One of these lineages (tMRCA = 0.510 [0.387-0.650] Ma) contains P. t. verus (tMRCA = 0.155 [0.101-0.213] Ma) and P. t. ellioti (formerly P. t. vellerosus; tMRCA = 0.157 [0.102-0.215] Ma), both of which are monophyletic. The other major lineage contains P. t. schweinfurthii (tMRCA = 0.111 [0.077-0.146] Ma), a monophyletic clade nested within the P. t. troglodytes lineage (tMRCA = 0.380 [0.296-0.476] Ma). We utilized two analysis techniques that may be of widespread interest. First, we implemented a Yule speciation prior across the entire primate tree with separate coalescent priors on each of the chimpanzee subspecies. The validity of this approach was confirmed by estimates based on more traditional techniques. We also suggest that accurate tMRCA estimates from large computationally difficult sequence alignments may be obtained by implementing our novel method of bootstrapping smaller randomly subsampled alignments.
Figures


References
-
- Bailey WJ, Hayasaka K, Skinner CG, Kehoe S, Sieu LC, Slightom JL, Goodman M. Reexamination of the African hominoid trichotomy with additional sequences from the primate beta-globin gene cluster. Mol Phylogenet Evol. 1992;1:97–135. - PubMed
-
- Ballard JW, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst. 2005;36:621–642.
-
- Booth AH. The Niger, the Volta and the Cahomey Gap as geographic barriers. Evolution. 1958;12:48–62.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

