The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor
- PMID: 20804771
- PMCID: PMC2949468
- DOI: 10.1016/j.jmb.2010.08.030
The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor
Abstract
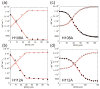
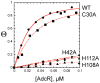
Streptococcus pneumoniae D39 AdcR (adhesin competence repressor) is the first metal-sensing member of the MarR (multiple antibiotic resistance repressor) family to be characterized. Expression profiling with a ΔadcR strain grown in liquid culture (brain-heart infusion) under microaerobic conditions revealed upregulation of 13 genes, including adcR and adcCBA, encoding a high-affinity ABC uptake system for zinc, and genes encoding cell-surface zinc-binding pneumococcal histidine triad (Pht) proteins and AdcAII (Lmb, laminin binding). The ΔadcR, H108Q and H112Q adcR mutant allelic strains grown in 0.2 mM Zn(II) exhibit a slow-growth phenotype and an approximately twofold increase in cell-associated Zn(II). Apo- and Zn(II)-bound AdcR are homodimers in solution and binding to a 28-mer DNA containing an adc operator is strongly stimulated by Zn(II) with K(DNA-Zn)=2.4 × 10(8) M(-1) (pH 6.0, 0.2 M NaCl, 25 °C). AdcR binds two Zn(II) per dimer, with stepwise Zn(II) affinities K(Zn1) and K(Zn2) of ≥10(9) M(-1) at pH 6.0 and ≥10(12) M(-1) at pH 8.0, and one to three lower affinity Zn(II) depending on the pH. X-ray absorption spectroscopy of the high-affinity site reveals a pentacoordinate N/O complex and no cysteine coordination, the latter finding corroborated by wild type-like functional properties of C30A AdcR. Alanine substitution of conserved residues His42 in the DNA-binding domain, and His108 and His112 in the C-terminal regulatory domain, abolish high-affinity Zn(II) binding and greatly reduce Zn(II)-activated binding to DNA. NMR studies reveal that these mutants adopt the same folded conformation as dimeric wild type apo-AdcR, but fail to conformationally switch upon Zn(II) binding. These studies implicate His42, His108 and H112 as metalloregulatory zinc ligands in S. pneumoniae AdcR.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures






Similar articles
-
Crystal structure of the zinc-dependent MarR family transcriptional regulator AdcR in the Zn(II)-bound state.J Am Chem Soc. 2011 Dec 14;133(49):19614-7. doi: 10.1021/ja2080532. Epub 2011 Nov 21. J Am Chem Soc. 2011. PMID: 22085181 Free PMC article.
-
Backbone and sterospecific methyl side chain resonance assignments of the homodimeric zinc sensor AdcR (32 kDa) in the apo- and Zn(II)-bound states.Biomol NMR Assign. 2014 Apr;8(1):11-4. doi: 10.1007/s12104-012-9442-6. Epub 2012 Nov 9. Biomol NMR Assign. 2014. PMID: 23138857 Free PMC article.
-
Physical characterization of the manganese-sensing pneumococcal surface antigen repressor from Streptococcus pneumoniae.Biochemistry. 2013 Oct 29;52(43):7689-701. doi: 10.1021/bi401132w. Epub 2013 Oct 14. Biochemistry. 2013. PMID: 24067066 Free PMC article.
-
Transcriptional response of Streptococcus pneumoniae to Zn2+) limitation and the repressor/activator function of AdcR.Metallomics. 2011 Jun;3(6):609-18. doi: 10.1039/c1mt00030f. Epub 2011 May 21. Metallomics. 2011. PMID: 21603707
-
Studying allosteric regulation in metal sensor proteins using computational methods.Adv Protein Chem Struct Biol. 2014;96:181-218. doi: 10.1016/bs.apcsb.2014.06.009. Epub 2014 Sep 6. Adv Protein Chem Struct Biol. 2014. PMID: 25443958 Review.
Cited by
-
Cell-free biosensors for rapid detection of water contaminants.Nat Biotechnol. 2020 Dec;38(12):1451-1459. doi: 10.1038/s41587-020-0571-7. Epub 2020 Jul 6. Nat Biotechnol. 2020. PMID: 32632301 Free PMC article.
-
Bacterial Strategies to Maintain Zinc Metallostasis at the Host-Pathogen Interface.J Biol Chem. 2016 Sep 30;291(40):20858-20868. doi: 10.1074/jbc.R116.742023. Epub 2016 Jul 26. J Biol Chem. 2016. PMID: 27462080 Free PMC article. Review.
-
Nutrient Zinc at the Host-Pathogen Interface.Trends Biochem Sci. 2019 Dec;44(12):1041-1056. doi: 10.1016/j.tibs.2019.06.010. Epub 2019 Jul 17. Trends Biochem Sci. 2019. PMID: 31326221 Free PMC article. Review.
-
Zinc is an inhibitor of the LdtR transcriptional activator.PLoS One. 2018 Apr 10;13(4):e0195746. doi: 10.1371/journal.pone.0195746. eCollection 2018. PLoS One. 2018. PMID: 29634775 Free PMC article.
-
Manganese acquisition and homeostasis at the host-pathogen interface.Front Cell Infect Microbiol. 2013 Dec 5;3:91. doi: 10.3389/fcimb.2013.00091. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24367765 Free PMC article. Review.
References
-
- Dintilhac A, Alloing G, Granadel C, Claverys JP. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. - PubMed
-
- Claverys JP, Dintilhac A, Mortier-Barriere I, Martin B, Alloing G. Regulation of competence for genetic transformation in Streptococcus pneumoniae. Soc Appl Bacteriol Symp Ser. 1997;26:32S–41S. - PubMed
-
- Dintilhac A, Claverys JP. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148:119–131. - PubMed
-
- Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

