The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage
- PMID: 20805324
- PMCID: PMC2935572
- DOI: 10.1083/jcb.200912135
The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage
Abstract
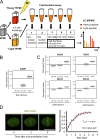
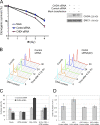
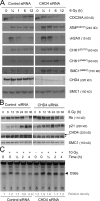
In response to ionizing radiation (IR), cells delay cell cycle progression and activate DNA repair. Both processes are vital for genome integrity, but the mechanisms involved in their coordination are not fully understood. In a mass spectrometry screen, we identified the adenosine triphosphate-dependent chromatin-remodeling protein CHD4 (chromodomain helicase DNA-binding protein 4) as a factor that becomes transiently immobilized on chromatin after IR. Knockdown of CHD4 triggers enhanced Cdc25A degradation and p21(Cip1) accumulation, which lead to more pronounced cyclin-dependent kinase inhibition and extended cell cycle delay. At DNA double-strand breaks, depletion of CHD4 disrupts the chromatin response at the level of the RNF168 ubiquitin ligase, which in turn impairs local ubiquitylation and BRCA1 assembly. These cell cycle and chromatin defects are accompanied by elevated spontaneous and IR-induced DNA breakage, reduced efficiency of DNA repair, and decreased clonogenic survival. Thus, CHD4 emerges as a novel genome caretaker and a factor that facilitates both checkpoint signaling and repair events after DNA damage.
Figures


References
-
- Bowen N.J., Fujita N., Kajita M., Wade P.A. 2004. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta. 1677:52–57 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

