Characterization of the CpxRA regulon in Haemophilus ducreyi
- PMID: 20805330
- PMCID: PMC2976327
- DOI: 10.1128/IAI.00678-10
Characterization of the CpxRA regulon in Haemophilus ducreyi
Abstract
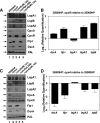
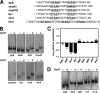
The Haemophilus ducreyi 35000HP genome encodes a homolog of the CpxRA two-component cell envelope stress response system originally characterized in Escherichia coli. CpxR, the cytoplasmic response regulator, was shown previously to be involved in repression of the expression of the lspB-lspA2 operon (M. Labandeira-Rey, J. R. Mock, and E. J. Hansen, Infect. Immun. 77:3402-3411, 2009). In the present study, the H. ducreyi CpxR and CpxA proteins were shown to closely resemble those of other well-studied bacterial species. A cpxA deletion mutant and a CpxR-overexpressing strain were used to explore the extent of the CpxRA regulon. DNA microarray and real-time reverse transcriptase (RT) PCR analyses indicated several potential regulatory targets for the H. ducreyi CpxRA two-component regulatory system. Electrophoretic mobility shift assays (EMSAs) were used to prove that H. ducreyi CpxR interacted with the promoter regions of genes encoding both known and putative virulence factors of H. ducreyi, including the lspB-lspA2 operon, the flp operon, and dsrA. Interestingly, the use of EMSAs also indicated that H. ducreyi CpxR did not bind to the promoter regions of several genes predicted to encode factors involved in the cell envelope stress response. Taken together, these data suggest that the CpxRA system in H. ducreyi, in contrast to that in E. coli, may be involved primarily in controlling expression of genes not involved in the cell envelope stress response.
Figures

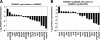
References
-
- Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. - PubMed
-
- Banks, K. E., K. R. Fortney, B. Baker, S. D. Billings, B. P. Katz, R. S. J. Munson, and S. M. Spinola. 2008. The enterobacterial common antigen-like gene cluster of Haemophilus ducreyi contributes to virulence in humans. J. Infect. Dis. 197:1531-1536. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

