The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression
- PMID: 20805357
- PMCID: PMC2953049
- DOI: 10.1128/MCB.00396-10
The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression
Abstract
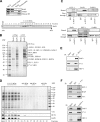
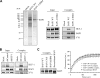
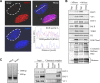
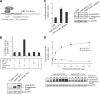
The candidate tumor suppressor BAP1 is a deubiquitinating enzyme (DUB) involved in the regulation of cell proliferation, although the molecular mechanisms governing its function remain poorly defined. BAP1 was recently shown to interact with and deubiquitinate the transcriptional regulator host cell factor 1 (HCF-1). Here we show that BAP1 assembles multiprotein complexes containing numerous transcription factors and cofactors, including HCF-1 and the transcription factor Yin Yang 1 (YY1). Through its coiled-coil motif, BAP1 directly interacts with the zinc fingers of YY1. Moreover, HCF-1 interacts with the middle region of YY1 encompassing the glycine-lysine-rich domain and is essential for the formation of a ternary complex with YY1 and BAP1 in vivo. BAP1 activates transcription in an enzymatic-activity-dependent manner and regulates the expression of a variety of genes involved in numerous cellular processes. We further show that BAP1 and HCF-1 are recruited by YY1 to the promoter of the cox7c gene, which encodes a mitochondrial protein used here as a model of BAP1-activated gene expression. Our findings (i) establish a direct link between BAP1 and the transcriptional control of genes regulating cell growth and proliferation and (ii) shed light on a novel mechanism of transcription regulation involving ubiquitin signaling.
Figures




References
-
- Calvano, S. E., W. Xiao, D. R. Richards, R. M. Felciano, H. V. Baker, R. J. Cho, R. O. Chen, B. H. Brownstein, J. P. Cobb, S. K. Tschoeke, C. Miller-Graziano, L. L. Moldawer, M. N. Mindrinos, R. W. Davis, R. G. Tompkins, and S. F. Lowry. 2005. A network-based analysis of systemic inflammation in humans. Nature 437:1032-1037. - PubMed
-
- Chen, Z. J., and L. J. Sun. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275-286. - PubMed
-
- Cho, Y. S., E. J. Kim, U. H. Park, H. S. Sin, and S. J. Um. 2006. Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J. Biol. Chem. 281:17588-17598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
