Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain
- PMID: 20805464
- PMCID: PMC2941324
- DOI: 10.1073/pnas.1007144107
Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain
Erratum in
- Proc Natl Acad Sci U S A. 2010 Sep 28;107(39):17059. Mas, Phillipe J [corrected to Mas, Philippe J]
Abstract
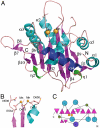
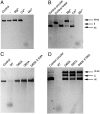
During viral replication, herpesviruses package their DNA into the procapsid by means of the terminase protein complex. In human cytomegalovirus (herpesvirus 5), the terminase is composed of subunits UL89 and UL56. UL89 cleaves the long DNA concatemers into unit-length genomes of appropriate length for encapsidation. We used ESPRIT, a high-throughput screening method, to identify a soluble purifiable fragment of UL89 from a library of 18,432 randomly truncated ul89 DNA constructs. The purified protein was crystallized and its three-dimensional structure was solved. This protein corresponds to the key nuclease domain of the terminase and shows an RNase H/integrase-like fold. We demonstrate that UL89-C has the capacity to process the DNA and that this function is dependent on Mn(2+) ions, two of which are located at the active site pocket. We also show that the nuclease function can be inactivated by raltegravir, a recently approved anti-AIDS drug that targets the HIV integrase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mocarski ES, Shenk T, Pass RF. In: Fields Virology. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2701–2772.
-
- Baines JD, Weller SK. In: Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Catalano CE, editor. New York: Springer; 2005. pp. 135–150.
-
- Bogner E. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev Med Virol. 2002;12:115–127. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

