Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK- anaplastic large-cell lymphoma
- PMID: 20805506
- PMCID: PMC2941277
- DOI: 10.1073/pnas.1009719107
Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK- anaplastic large-cell lymphoma
Abstract
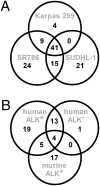
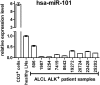
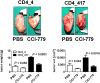
Aberrant anaplastic lymphoma kinase (ALK) expression is a defining feature of many human cancers and was identified first in anaplastic large-cell lymphoma (ALCL), an aggressive non-Hodgkin T-cell lymphoma. Since that time, many studies have set out to identify the mechanisms used by aberrant ALK toward tumorigenesis. We have identified a distinct profile of micro-RNAs (miRNAs) that characterize ALCL; furthermore, this profile distinguishes ALK(+) from ALK(-) subtypes, and thus points toward potential mechanisms of tumorigenesis induced by aberrant ALK. Using a nucleophosmin-ALK transgenic mouse model as well as human primary ALCL tumor tissues and human ALCL-derived cell lines, we reveal a set of overlapping deregulated miRNAs that might be implicated in the development and progression of ALCL. Importantly, ALK(+) and ALK(-) ALCL could be distinguished by a distinct profile of "oncomirs": Five members of the miR-17-92 cluster were expressed more highly in ALK(+) ALCL, whereas miR-155 was expressed more than 10-fold higher in ALK(-) ALCL. Moreover, miR-101 was down-regulated in all ALCL model systems, but its forced expression attenuated cell proliferation only in ALK(+) and not in ALK(-) cell lines, perhaps suggesting different modes of ALK-dependent regulation of its target proteins. Furthermore, inhibition of mTOR, which is targeted by miR-101, led to reduced tumor growth in engrafted ALCL mouse models. In addition to future therapeutical and diagnostic applications, it will be of interest to study the physiological implications and prognostic value of the identified miRNA profiles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Benz-Lemoine E, et al. Malignant histiocytosis: A specific t(2;5)(p23;q35) translocation? Review of the literature. Blood. 1988;72:1045–1047. - PubMed
-
- Fischer P, et al. A Ki-1 (CD30)-positive human cell line (Karpas 299) established from a high-grade non-Hodgkin's lymphoma, showing a 2;5 translocation and rearrangement of the T-cell receptor beta-chain gene. Blood. 1988;72:234–240. - PubMed
-
- Mason DY, et al. CD30-positive large cell lymphomas (‘Ki-1 lymphoma’) are associated with a chromosomal translocation involving 5q35. Br J Haematol. 1990;74:161–168. - PubMed
-
- Morris SW, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. - PubMed
-
- Lim MS, et al. The proteomic signature of NPM/ALK reveals deregulation of multiple cellular pathways. Blood. 2009;114:1585–1595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

