Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase
- PMID: 20805881
- PMCID: PMC2923608
- DOI: 10.1371/journal.pone.0012245
Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase
Abstract
Mycobacterium tuberculosis DNA gyrase, an indispensable nanomachine involved in the regulation of DNA topology, is the only type II topoisomerase present in this organism and is hence the sole target for quinolone action, a crucial drug active against multidrug-resistant tuberculosis. To understand at an atomic level the quinolone resistance mechanism, which emerges in extensively drug resistant tuberculosis, we performed combined functional, biophysical and structural studies of the two individual domains constituting the catalytic DNA gyrase reaction core, namely the Toprim and the breakage-reunion domains. This allowed us to produce a model of the catalytic reaction core in complex with DNA and a quinolone molecule, identifying original mechanistic properties of quinolone binding and clarifying the relationships between amino acid mutations and resistance phenotype of M. tuberculosis DNA gyrase. These results are compatible with our previous studies on quinolone resistance. Interestingly, the structure of the entire breakage-reunion domain revealed a new interaction, in which the Quinolone-Binding Pocket (QBP) is blocked by the N-terminal helix of a symmetry-related molecule. This interaction provides useful starting points for designing peptide based inhibitors that target DNA gyrase to prevent its binding to DNA.
Conflict of interest statement
Figures

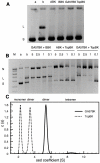

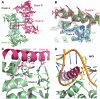



References
-
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. - PubMed
-
- Gadelle D, Filee J, Buhler C, Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;25:232–242. - PubMed
-
- Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem Soc Trans. 2005;33:1465–1470. - PubMed
-
- Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

