Gut microbial gene expression in mother-fed and formula-fed piglets
- PMID: 20805981
- PMCID: PMC2929194
- DOI: 10.1371/journal.pone.0012459
Gut microbial gene expression in mother-fed and formula-fed piglets
Abstract
Background: Effects of diet on the structure and function of gut microbial communities in newborn infants are poorly understood. High-resolution molecular studies are needed to definitively ascertain whether gut microbial communities are distinct in milk-fed and formula-fed infants.
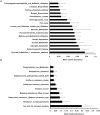
Methodology/principal findings: Pyrosequencing-based whole transcriptome shotgun sequencing (RNA-seq) was used to evaluate community wide gut microbial gene expression in 21 day old neonatal piglets fed either with sow's milk (mother fed, MF; n = 4) or with artificial formula (formula fed, FF; n = 4). Microbial DNA and RNA were harvested from cecal contents for each animal. cDNA libraries and 16S rDNA amplicons were sequenced on the Roche 454 GS-FLX Titanium system. Communities were similar at the level of phylum but were dissimilar at the level of genus; Prevotella was the dominant genus within MF samples and Bacteroides was most abundant within FF samples. Screened cDNA sequences were assigned functional annotations by the MG-RAST annotation pipeline and based upon best-BLASTX-hits to the NCBI COG database. Patterns of gene expression were very similar in MF and FF animals. All samples were enriched with transcripts encoding enzymes for carbohydrate and protein metabolism, as well as proteins involved in stress response, binding to host epithelium, and lipopolysaccharide metabolism. Carbohydrate utilization transcripts were generally similar in both groups. The abundance of enzymes involved in several pathways related to amino acid metabolism (e.g., arginine metabolism) and oxidative stress response differed in MF and FF animals.
Conclusions/significance: Abundant transcripts identified in this study likely contribute to a core microbial metatranscriptome in the distal intestine. Although microbial community gene expression was generally similar in the cecal contents of MF and FF neonatal piglets, several differentially abundant gene clusters were identified. Further investigations of gut microbial gene expression will contribute to a better understanding of normal and abnormal enteric microbiology in animals and humans.
Conflict of interest statement
Figures

Similar articles
-
Murine gut microbiota and transcriptome are diet dependent.Ann Surg. 2013 Feb;257(2):287-94. doi: 10.1097/SLA.0b013e318262a6a6. Ann Surg. 2013. PMID: 23001074
-
Bacterial and Fungal Adaptations in Cecum and Distal Colon of Piglets Fed With Dairy-Based Milk Formula in Comparison With Human Milk.Front Microbiol. 2022 Mar 23;13:801854. doi: 10.3389/fmicb.2022.801854. eCollection 2022. Front Microbiol. 2022. PMID: 35401465 Free PMC article.
-
Milk Formula Diet Alters Bacterial and Host Protein Profile in Comparison to Human Milk Diet in Neonatal Piglet Model.Nutrients. 2021 Oct 22;13(11):3718. doi: 10.3390/nu13113718. Nutrients. 2021. PMID: 34835974 Free PMC article.
-
Neonatal Diet Impacts Bioregional Microbiota Composition in Piglets Fed Human Breast Milk or Infant Formula.J Nutr. 2019 Dec 1;149(12):2236-2246. doi: 10.1093/jn/nxz170. J Nutr. 2019. PMID: 31373372 Free PMC article.
-
Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides.Adv Nutr. 2012 May 1;3(3):450S-5S. doi: 10.3945/an.112.001859. Adv Nutr. 2012. PMID: 22585924 Free PMC article. Review.
Cited by
-
High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days.Appl Environ Microbiol. 2012 Jun;78(12):4217-24. doi: 10.1128/AEM.00307-12. Epub 2012 Mar 30. Appl Environ Microbiol. 2012. PMID: 22467509 Free PMC article.
-
The effect of feeding Bt MON810 maize to pigs for 110 days on intestinal microbiota.PLoS One. 2012;7(5):e33668. doi: 10.1371/journal.pone.0033668. Epub 2012 May 4. PLoS One. 2012. PMID: 22574106 Free PMC article.
-
Maternal imprinting of the neonatal microbiota colonization in intrauterine growth restricted piglets: a review.J Anim Sci Biotechnol. 2019 Nov 11;10:88. doi: 10.1186/s40104-019-0397-7. eCollection 2019. J Anim Sci Biotechnol. 2019. PMID: 31737268 Free PMC article. Review.
-
Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age.J Microbiol. 2018 May;56(5):346-355. doi: 10.1007/s12275-018-7486-8. Epub 2018 May 2. J Microbiol. 2018. PMID: 29721832
-
A Nutrigenomics Approach Using RNA Sequencing Technology to Study Nutrient-Gene Interactions in Agricultural Animals.Curr Dev Nutr. 2019 Jul 15;3(8):nzz082. doi: 10.1093/cdn/nzz082. eCollection 2019 Aug. Curr Dev Nutr. 2019. PMID: 31414073 Free PMC article. Review.
References
-
- Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–45S. - PubMed
-
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. - PubMed
-
- Lundequist B, Nord CE, Winberg J. The composition of the faecal microflora in breastfed and bottle fed infants from birth to eight weeks. Acta Paediatr Scand 1985. 1985;74:45–51. - PubMed
-
- Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol. 2005;54:77–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

