In vitro transformation of primary human CD34+ cells by AML fusion oncogenes: early gene expression profiling reveals possible drug target in AML
- PMID: 20805992
- PMCID: PMC2929205
- DOI: 10.1371/journal.pone.0012464
In vitro transformation of primary human CD34+ cells by AML fusion oncogenes: early gene expression profiling reveals possible drug target in AML
Abstract
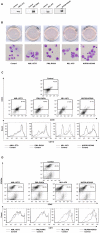
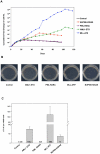
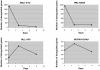
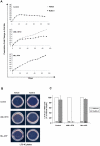
Different fusion oncogenes in acute myeloid leukemia (AML) have distinct clinical and laboratory features suggesting different modes of malignant transformation. Here we compare the in vitro effects of representatives of 4 major groups of AML fusion oncogenes on primary human CD34+ cells. As expected from their clinical similarities, MLL-AF9 and NUP98-HOXA9 had very similar effects in vitro. They both caused erythroid hyperplasia and a clear block in erythroid and myeloid maturation. On the other hand, AML1-ETO and PML-RARA had only modest effects on myeloid and erythroid differentiation. All oncogenes except PML-RARA caused a dramatic increase in long-term proliferation and self-renewal. Gene expression profiling revealed two distinct temporal patterns of gene deregulation. Gene deregulation by MLL-AF9 and NUP98-HOXA9 peaked 3 days after transduction. In contrast, the vast majority of gene deregulation by AML1-ETO and PML-RARA occurred within 6 hours, followed by a dramatic drop in the numbers of deregulated genes. Interestingly, the p53 inhibitor MDM2 was upregulated by AML1-ETO at 6 hours. Nutlin-3, an inhibitor of the interaction between MDM2 and p53, specifically inhibited the proliferation and self-renewal of primary human CD34+ cells transduced with AML1-ETO, suggesting that MDM2 upregulation plays a role in cell transformation by AML1-ETO. These data show that differences among AML fusion oncogenes can be recapitulated in vitro using primary human CD34+ cells and that early gene expression profiling in these cells can reveal potential drug targets in AML.
Conflict of interest statement
Figures

Similar articles
-
AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1.Blood. 2006 Apr 15;107(8):3303-12. doi: 10.1182/blood-2005-04-1656. Epub 2005 Dec 27. Blood. 2006. PMID: 16380455
-
Interaction of c-Myb with p300 is required for the induction of acute myeloid leukemia (AML) by human AML oncogenes.Blood. 2014 Apr 24;123(17):2682-90. doi: 10.1182/blood-2012-02-413187. Epub 2014 Mar 4. Blood. 2014. PMID: 24596419
-
Relationships between AML1-ETO and MLL-AF9 fusion gene expressions and hematological parameters in acute myeloid leukemia.Gulf J Oncolog. 2020 Sep;1(34):65-69. Gulf J Oncolog. 2020. PMID: 33431365
-
Molecular pathways mediating MDS/AML with focus on AML1/RUNX1 point mutations.J Cell Physiol. 2009 Jul;220(1):16-20. doi: 10.1002/jcp.21769. J Cell Physiol. 2009. PMID: 19334039 Review.
-
The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias.Haematologica. 1997 May-Jun;82(3):364-70. Haematologica. 1997. PMID: 9234595 Review.
Cited by
-
ZNF521 sustains the differentiation block in MLL-rearranged acute myeloid leukemia.Oncotarget. 2017 Apr 18;8(16):26129-26141. doi: 10.18632/oncotarget.15387. Oncotarget. 2017. PMID: 28412727 Free PMC article.
-
Turning Stem Cells Bad: Generation of Clinically Relevant Models of Human Acute Myeloid Leukemia through Gene Delivery- or Genome Editing-Based Approaches.Molecules. 2018 Aug 17;23(8):2060. doi: 10.3390/molecules23082060. Molecules. 2018. PMID: 30126100 Free PMC article. Review.
-
Engineering an inducible leukemia-associated fusion protein enables large-scale ex vivo production of functional human phagocytes.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2312499121. doi: 10.1073/pnas.2312499121. Epub 2024 Jun 10. Proc Natl Acad Sci U S A. 2024. PMID: 38857395 Free PMC article.
-
Oncogenic fusion proteins expressed in immature hematopoietic cells fail to recapitulate the transcriptional changes observed in human AML.Oncogenesis. 2014 Jun 16;3(6):e106. doi: 10.1038/oncsis.2014.22. Oncogenesis. 2014. PMID: 24932908 Free PMC article.
-
NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis.Cancer Cell. 2016 Dec 12;30(6):863-878. doi: 10.1016/j.ccell.2016.10.019. Epub 2016 Nov 23. Cancer Cell. 2016. PMID: 27889185 Free PMC article.
References
-
- Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. - PubMed
-
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. - PubMed
-
- Vitoux D, Nasr R, de The H. Acute promyelocytic leukemia: new issues on pathogenesis and treatment response. Int J Biochem Cell Biol. 2007;39:1063–1070. - PubMed
-
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia 2009 - PubMed
-
- Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, et al. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–2402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

