Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing
- PMID: 20807199
- PMCID: PMC2959132
- DOI: 10.1111/j.1365-2958.2010.07351.x
Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing
Abstract
The UPF0054 protein family is highly conserved with homologues present in nearly every sequenced bacterium. In some bacteria, the respective gene is essential, while in others its loss results in a highly pleiotropic phenotype. Despite detailed structural studies, a cellular role for this protein family has remained unknown. We report here that deletion of the Escherichia coli homologue, YbeY, causes striking defects that affect ribosome activity, translational fidelity and ribosome assembly. Mapping of 16S, 23S and 5S rRNA termini reveals that YbeY influences the maturation of all three rRNAs, with a particularly strong effect on maturation at both the 5'- and 3'-ends of 16S rRNA as well as maturation of the 5'-termini of 23S and 5S rRNAs. Furthermore, we demonstrate strong genetic interactions between ybeY and rnc (encoding RNase III), ybeY and rnr (encoding RNase R), and ybeY and pnp (encoding PNPase), further suggesting a role for YbeY in rRNA maturation. Mutation of highly conserved amino acids in YbeY, allowed the identification of two residues (H114, R59) that were found to have a significant effect in vivo. We discuss the implications of these findings for rRNA maturation and ribosome assembly in bacteria.
© 2010 Blackwell Publishing Ltd.
Figures


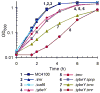

Similar articles
-
Elevated Levels of Era GTPase Improve Growth, 16S rRNA Processing, and 70S Ribosome Assembly of Escherichia coli Lacking Highly Conserved Multifunctional YbeY Endoribonuclease.J Bacteriol. 2018 Aug 10;200(17):e00278-18. doi: 10.1128/JB.00278-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29914987 Free PMC article.
-
Identification of YbeY-Protein Interactions Involved in 16S rRNA Maturation and Stress Regulation in Escherichia coli.mBio. 2016 Nov 8;7(6):e01785-16. doi: 10.1128/mBio.01785-16. mBio. 2016. PMID: 27834201 Free PMC article.
-
Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli.J Biol Chem. 2018 Jun 8;293(23):9006-9016. doi: 10.1074/jbc.RA117.000300. Epub 2018 Apr 20. J Biol Chem. 2018. PMID: 29678883 Free PMC article.
-
What do we know about ribosomal RNA methylation in Escherichia coli?Biochimie. 2015 Oct;117:110-8. doi: 10.1016/j.biochi.2014.11.019. Epub 2014 Dec 13. Biochimie. 2015. PMID: 25511423 Review.
-
Intermolecular base-paired interaction between complementary sequences present near the 3' ends of 5S rRNA and 18S (16S) rRNA might be involved in the reversible association of ribosomal subunits.Nucleic Acids Res. 1979 Dec 11;7(7):1913-29. doi: 10.1093/nar/7.7.1913. Nucleic Acids Res. 1979. PMID: 94160 Free PMC article. Review.
Cited by
-
The Escherichia coli translation-associated heat shock protein YbeY is involved in rRNA transcription antitermination.PLoS One. 2013 Apr 29;8(4):e62297. doi: 10.1371/journal.pone.0062297. Print 2013. PLoS One. 2013. PMID: 23638028 Free PMC article.
-
GTPase Era at the heart of ribosome assembly.Front Mol Biosci. 2023 Oct 4;10:1263433. doi: 10.3389/fmolb.2023.1263433. eCollection 2023. Front Mol Biosci. 2023. PMID: 37860580 Free PMC article. Review.
-
Elevated Levels of Era GTPase Improve Growth, 16S rRNA Processing, and 70S Ribosome Assembly of Escherichia coli Lacking Highly Conserved Multifunctional YbeY Endoribonuclease.J Bacteriol. 2018 Aug 10;200(17):e00278-18. doi: 10.1128/JB.00278-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29914987 Free PMC article.
-
Identification of YbeY-Protein Interactions Involved in 16S rRNA Maturation and Stress Regulation in Escherichia coli.mBio. 2016 Nov 8;7(6):e01785-16. doi: 10.1128/mBio.01785-16. mBio. 2016. PMID: 27834201 Free PMC article.
-
YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa.Appl Environ Microbiol. 2021 Mar 1;87(5):e02171-20. doi: 10.1128/AEM.02171-20. Epub 2020 Dec 11. Appl Environ Microbiol. 2021. PMID: 33310711 Free PMC article.
References
-
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H. Largescale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

