Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition
- PMID: 20807527
- PMCID: PMC3142750
- DOI: 10.1016/j.ydbio.2010.08.020
Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition
Abstract
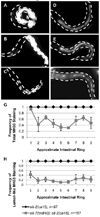
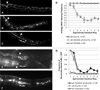
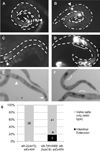
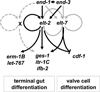
The transition from specification of cell identity to the differentiation of cells into an appropriate and enduring state is critical to the development of embryos. Transcriptional profiling in Caenorhabditis elegans has revealed a large number of genes that are expressed in the fully differentiated intestine; however, no regulatory factor has been found to be essential to initiate their expression once the endoderm has been specified. These gut-expressed genes possess a preponderance of GATA factor binding sites and one GATA factor, ELT-2, fulfills the expected characteristics of a key regulator of these genes based on its persistent expression exclusively in the developing and differentiated intestine and its ability to bind these regulatory sites. However, a striking characteristic of elt-2(0) knockout mutants is that while they die shortly after hatching owing to an obstructed gut passage, they nevertheless contain a gut that has undergone complete morphological differentiation. We have discovered a second gut-specific GATA factor, ELT-7, that profoundly synergizes with ELT-2 to create a transcriptional switch essential for gut cell differentiation. ELT-7 is first expressed in the early endoderm lineage and, when expressed ectopically, is sufficient to activate gut differentiation in nonendodermal progenitors. elt-7 is transcriptionally activated by the redundant endoderm-specifying factors END-1 and -3, and its product in turn activates both its own expression and that of elt-2, constituting an apparent positive feedback system. While elt-7 loss-of-function mutants lack a discernible phenotype, simultaneous loss of both elt-7 and elt-2 results in a striking all-or-none block to morphological differentiation of groups of gut cells with a region-specific bias, as well as reduced or abolished gut-specific expression of a number of terminal differentiation genes. ELT-2 and -7 synergize not only in activation of gene expression but also in repression of a gene that is normally expressed in the valve cells, which immediately flank the termini of the gut tube. Our results point to a developmental strategy whereby positive feedback and cross-regulatory interactions between two synergistically acting regulatory factors promote a decisive and persistent transition of specified endoderm progenitors into the program of intestinal differentiation.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
The function and regulation of the GATA factor ELT-2 in the C. elegans endoderm.Development. 2016 Feb 1;143(3):483-91. doi: 10.1242/dev.130914. Epub 2015 Dec 23. Development. 2016. PMID: 26700680 Free PMC article.
-
A Strategy To Isolate Modifiers of Caenorhabditis elegans Lethal Mutations: Investigating the Endoderm Specifying Ability of the Intestinal Differentiation GATA Factor ELT-2.G3 (Bethesda). 2018 May 4;8(5):1425-1437. doi: 10.1534/g3.118.200079. G3 (Bethesda). 2018. PMID: 29593072 Free PMC article.
-
Partially compromised specification causes stochastic effects on gut development in C. elegans.Dev Biol. 2017 Jul 1;427(1):49-60. doi: 10.1016/j.ydbio.2017.05.007. Epub 2017 May 11. Dev Biol. 2017. PMID: 28502614
-
The Caenorhabditis elegans intestine.Wiley Interdiscip Rev Dev Biol. 2013 May-Jun;2(3):347-67. doi: 10.1002/wdev.93. Epub 2012 Oct 9. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799580 Review.
-
GATA factors as key regulatory molecules in the development of Drosophila endoderm.Dev Growth Differ. 2005 Dec;47(9):581-9. doi: 10.1111/j.1440-169X.2005.00836.x. Dev Growth Differ. 2005. PMID: 16316403 Review.
Cited by
-
Deactivation of the GATA Transcription Factor ELT-2 Is a Major Driver of Normal Aging in C. elegans.PLoS Genet. 2016 Apr 12;12(4):e1005956. doi: 10.1371/journal.pgen.1005956. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27070429 Free PMC article.
-
Transorganogenesis and transdifferentiation in C. elegans are dependent on differentiated cell identity.Dev Biol. 2016 Dec 1;420(1):136-147. doi: 10.1016/j.ydbio.2016.09.020. Epub 2016 Oct 4. Dev Biol. 2016. PMID: 27717645 Free PMC article.
-
Feedforward regulatory logic controls the specification-to-differentiation transition and terminal cell fate during Caenorhabditis elegans endoderm development.Development. 2022 Jun 15;149(12):dev200337. doi: 10.1242/dev.200337. Epub 2022 Jun 27. Development. 2022. PMID: 35758255 Free PMC article.
-
Evolutionary Change in Gut Specification in Caenorhabditis Centers on the GATA Factor ELT-3 in an Example of Developmental System Drift.J Dev Biol. 2023 Jul 8;11(3):32. doi: 10.3390/jdb11030032. J Dev Biol. 2023. PMID: 37489333 Free PMC article. Review.
-
Evolution of Developmental GATA Factors in Nematodes.J Dev Biol. 2020 Nov 16;8(4):27. doi: 10.3390/jdb8040027. J Dev Biol. 2020. PMID: 33207804 Free PMC article.
References
-
- Ahringer J. In: Reverse genetics. Wormbook, editor. WormBook: The C. elegans Research Community; 2006. Apr 6,
-
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. - PubMed
-
- Beh CT, Ferrari DC, Chung MA, McGhee JD. An acid phosphatase as a biochemical marker for intestinal development in the nematode Caenorhabditis elegans. Dev Biol. 1991;147:133–143. - PubMed
-
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–125. - PubMed
-
- Bossinger O, Fukushige T, Claeys M, Borgonie G, McGhee JD. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev Biol. 2004;268:448–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

