Mechanism of phosphorylation-induced activation of phospholipase C-gamma isozymes
- PMID: 20807769
- PMCID: PMC2975207
- DOI: 10.1074/jbc.M110.166512
Mechanism of phosphorylation-induced activation of phospholipase C-gamma isozymes
Abstract
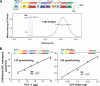
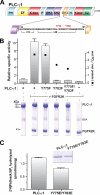
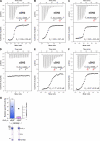
The lipase activity of most phospholipases C (PLCs) is basally repressed by a highly degenerate and mostly disordered X/Y linker inserted within the catalytic domain. Release of this auto-inhibition is driven by electrostatic repulsion between the plasma membrane and the electronegative X/Y linker. In contrast, PLC-γ isozymes (PLC-γ1 and -γ2) are structurally distinct from other PLCs because multiple domains are present in their X/Y linker. Moreover, although many tyrosine kinases directly phosphorylate PLC-γ isozymes to enhance their lipase activity, the underlying molecular mechanism of this activation remains unclear. Here we define the mechanism for the unique regulation of PLC-γ isozymes by their X/Y linker. Specifically, we identify the C-terminal SH2 domain within the X/Y linker as the critical determinant for auto-inhibition. Tyrosine phosphorylation of the X/Y linker mediates high affinity intramolecular interaction with the C-terminal SH2 domain that is coupled to a large conformational rearrangement and release of auto-inhibition. Consequently, PLC-γ isozymes link phosphorylation to phospholipase activation by elaborating upon primordial regulatory mechanisms found in other PLCs.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

