Identification of an endothelin-converting enzyme-2-specific fluorigenic substrate and development of an in vitro and ex vivo enzymatic assay
- PMID: 20807771
- PMCID: PMC2966053
- DOI: 10.1074/jbc.M110.120576
Identification of an endothelin-converting enzyme-2-specific fluorigenic substrate and development of an in vitro and ex vivo enzymatic assay
Abstract
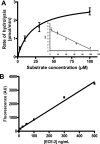
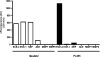
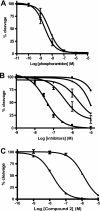
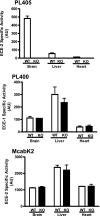
Endothelin-converting enzyme-2 (ECE-2) is a membrane-bound zinc-dependent metalloprotease that shares a high degree of sequence homology with ECE-1, but displays an acidic pH optimum characteristic of maturing enzymes acting late in the secretory pathway. Although ECE-2, like ECE-1, can cleave the big endothelin intermediate to produce the vasoconstrictive endothelin peptide, its true physiological function remains to be elucidated, a task that is hampered by the lack of specific tools to study and discriminate ECE-2 from ECE-1, i.e. specific substrates and/or specific inhibitors. To fill this gap, we searched for novel ECE-specific peptide substrates. To this end, peptides derived from the big endothelin intermediate were tested using ECE-1 and ECE-2, leading to the identification of an ECE-1-specific substrate. Moreover, screening of our proprietary fluorigenic peptide Fluofast® libraries using ECE-1 and ECE-2 allowed the identification of Ac-SKG-Pya-F-W-Nop-GGK-NH(2) (PL405), as a specific and high affinity ECE-2 substrate. Indeed, ECE-2 cleaved PL405 at the Pya-F amide bond with a specificity constant (k(cat)/K(m)) of 8.1 ± 0.9 × 10(3) M(-1) s(-1). Using this novel substrate, we also characterized the first potent (K(i) = 7.7 ± 0.3 nM) and relatively selective ECE-2 inhibitor and developed a quantitative fluorigenic ECE-2 assay. The assay was used to study the ex vivo ECE-2 activity in wild type and ECE-2 knock-out tissues and was found to truly reflect ECE-2 expression patterns. The PL405 assay is thus the first tool to study ECE-2 inhibition using high throughput screening or for ex vivo ECE-2 quantification.
Figures


Similar articles
-
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH optimum.J Biol Chem. 1995 Jun 23;270(25):15262-8. doi: 10.1074/jbc.270.25.15262. J Biol Chem. 1995. PMID: 7797512
-
Disulfide bonds in big ET-1 are essential for the specific cleavage at the Trp(21)-Val(22) bond by soluble endothelin converting enzyme-1 from baculovirus/insect cells.Arch Biochem Biophys. 2000 Jan 15;373(2):385-93. doi: 10.1006/abbi.1999.1586. Arch Biochem Biophys. 2000. PMID: 10620363
-
Highly sensitive and selective fluorescence assays for rapid screening of endothelin-converting enzyme inhibitors.Biochem J. 2001 Jun 15;356(Pt 3):813-9. doi: 10.1042/0264-6021:3560813. Biochem J. 2001. PMID: 11389689 Free PMC article.
-
Molecular pharmacology of endothelin converting enzymes.Biochem Pharmacol. 1996 Jan 26;51(2):91-102. doi: 10.1016/0006-2952(95)02036-5. Biochem Pharmacol. 1996. PMID: 8615890 Review.
-
The endothelin system and endothelin-converting enzyme in the brain: molecular and cellular studies.Neurochem Res. 1997 Aug;22(8):1033-40. doi: 10.1023/a:1022435111928. Neurochem Res. 1997. PMID: 9239759 Review.
Cited by
-
Endothelin-converting enzyme 2 differentially regulates opioid receptor activity.Br J Pharmacol. 2015 Jan;172(2):704-19. doi: 10.1111/bph.12833. Epub 2014 Sep 5. Br J Pharmacol. 2015. PMID: 24990314 Free PMC article.
-
Opioid receptor function is regulated by post-endocytic peptide processing.J Biol Chem. 2014 Jul 11;289(28):19613-26. doi: 10.1074/jbc.M113.537704. Epub 2014 May 20. J Biol Chem. 2014. PMID: 24847082 Free PMC article.
-
Identification of Hypoxia-Specific Biomarkers in Salmonids Using RNA-Sequencing and Validation Using High-Throughput qPCR.G3 (Bethesda). 2020 Sep 2;10(9):3321-3336. doi: 10.1534/g3.120.401487. G3 (Bethesda). 2020. PMID: 32694198 Free PMC article.
-
Neprilysin Inhibitors and Bradykinin.Front Med (Lausanne). 2018 Sep 19;5:257. doi: 10.3389/fmed.2018.00257. eCollection 2018. Front Med (Lausanne). 2018. PMID: 30283782 Free PMC article. Review.
References
-
- Xu D., Emoto N., Giaid A., Slaughter C., Kaw S., deWit D., Yanagisawa M. (1994) Cell 78, 473–485 - PubMed
-
- Emoto N., Yanagisawa M. (1995) J. Biol. Chem. 270, 15262–15268 - PubMed
-
- Roques B. P., Noble F., Daugé V., Fournié-Zaluski M. C., Beaumont A. (1993) Pharmacol Rev. 45, 87–146 - PubMed
-
- Valdenaire O., Richards J. G., Faull R. L., Schweizer A. (1999) Brain Res. Mol. Brain Res. 64, 211–221 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

