A multiprotein complex necessary for both transcription and DNA replication at the β-globin locus
- PMID: 20808282
- PMCID: PMC2957209
- DOI: 10.1038/emboj.2010.204
A multiprotein complex necessary for both transcription and DNA replication at the β-globin locus
Abstract
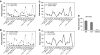
DNA replication, repair, transcription and chromatin structure are intricately associated nuclear processes, but the molecular links between these events are often obscure. In this study, we have surveyed the protein complexes that bind at β-globin locus control region, and purified and characterized the function of one such multiprotein complex from human erythroleukemic K562 cells. We further validated the existence of this complex in human CD34+ cell-derived normal erythroid cells. This complex contains ILF2/ILF3 transcription factors, p300 acetyltransferase and proteins associated with DNA replication, transcription and repair. RNAi knockdown of ILF2, a DNA-binding component of this complex, abrogates the recruitment of the complex to its cognate DNA sequence and inhibits transcription, histone acetylation and usage of the origin of DNA replication at the β-globin locus. These results imply a direct link between mammalian DNA replication, transcription and histone acetylation mediated by a single multiprotein complex.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
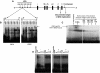

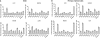


Similar articles
-
Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation.Blood. 2010 Sep 30;116(13):2356-64. doi: 10.1182/blood-2010-03-272252. Epub 2010 Jun 22. Blood. 2010. PMID: 20570862 Free PMC article.
-
Histone acetylation contributes to chromatin looping between the locus control region and globin gene by influencing hypersensitive site formation.Biochim Biophys Acta. 2013 Sep;1829(9):963-9. doi: 10.1016/j.bbagrm.2013.04.006. Epub 2013 Apr 20. Biochim Biophys Acta. 2013. PMID: 23607989
-
Identification of KAP-1-associated complexes negatively regulating the Ey and β-major globin genes in the β-globin locus.J Proteomics. 2013 Mar 27;80:132-44. doi: 10.1016/j.jprot.2012.12.014. Epub 2013 Jan 2. J Proteomics. 2013. PMID: 23291531
-
The mammalian beta globin origin of DNA replication.Front Biosci. 2004 Sep 1;9:2540-7. doi: 10.2741/1415. Front Biosci. 2004. PMID: 15358579 Review.
-
Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology.Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub 2016 Jan 26. Biochemistry. 2016. PMID: 26745824 Review.
Cited by
-
The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining.Mol Cell Biol. 2011 Dec;31(23):4832-43. doi: 10.1128/MCB.05849-11. Epub 2011 Oct 3. Mol Cell Biol. 2011. PMID: 21969602 Free PMC article.
-
A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells.Nat Commun. 2016 Jun 8;7:11748. doi: 10.1038/ncomms11748. Nat Commun. 2016. PMID: 27272143 Free PMC article.
-
Looping back to leap forward: transcription enters a new era.Cell. 2014 Mar 27;157(1):13-25. doi: 10.1016/j.cell.2014.02.009. Cell. 2014. PMID: 24679523 Free PMC article. Review.
-
Negative regulation of HBG1/2 expression through S6K by long noncoding RNA NR_120526.Transl Pediatr. 2023 May 30;12(5):907-917. doi: 10.21037/tp-23-174. Epub 2023 May 24. Transl Pediatr. 2023. PMID: 37305725 Free PMC article.
-
The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses.Biochim Biophys Acta. 2012 Jul;1819(7):794-801. doi: 10.1016/j.bbagrm.2012.01.017. Epub 2012 Feb 8. Biochim Biophys Acta. 2012. PMID: 22342530 Free PMC article. Review.
References
-
- Aladjem MI (2004) The mammalian beta globin origin of DNA replication. Front Biosci 9: 2540–2547 - PubMed
-
- Bridges BA (2005) Error-prone DNA repair and translesion synthesis: focus on the replication fork. DNA Repair (Amst) 4: 618–619, 634 - PubMed
-
- Chen J, Kremer CS, Bender TP (2006) The carbonic anhydrase I locus contains a c-Myb target promoter and modulates differentiation of murine erythroleukemia cells. Oncogene 25: 2758–2772 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

