Dcas supports cell polarization and cell-cell adhesion complexes in development
- PMID: 20808771
- PMCID: PMC2927436
- DOI: 10.1371/journal.pone.0012369
Dcas supports cell polarization and cell-cell adhesion complexes in development
Abstract
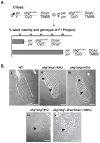
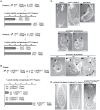
Mammalian Cas proteins regulate cell migration, division and survival, and are often deregulated in cancer. However, the presence of four paralogous Cas family members in mammals (BCAR1/p130Cas, EFS/Sin1, NEDD9/HEF1/Cas-L, and CASS4/HEPL) has limited their analysis in development. We deleted the single Drosophila Cas gene, Dcas, to probe the developmental function of Dcas. Loss of Dcas had limited effect on embryonal development. However, we found that Dcas is an important modulator of the severity of the developmental phenotypes of mutations affecting integrins (If and mew) and their downstream effectors Fak56D or Src42A. Strikingly, embryonic lethal Fak56D-Dcas double mutant embryos had extensive cell polarity defects, including mislocalization and reduced expression of E-cadherin. Further genetic analysis established that loss of Dcas modified the embryonal lethal phenotypes of embryos with mutations in E-cadherin (Shg) or its signaling partners p120- and beta-catenin (Arm). These results support an important role for Cas proteins in cell-cell adhesion signaling in development.
Conflict of interest statement
Figures





Similar articles
-
NEDD9 and BCAR1 negatively regulate E-cadherin membrane localization, and promote E-cadherin degradation.PLoS One. 2011;6(7):e22102. doi: 10.1371/journal.pone.0022102. Epub 2011 Jul 12. PLoS One. 2011. PMID: 21765937 Free PMC article.
-
α-Catenin stabilises Cadherin-Catenin complexes and modulates actomyosin dynamics to allow pulsatile apical contraction.J Cell Sci. 2016 Dec 15;129(24):4496-4508. doi: 10.1242/jcs.193268. Epub 2016 Nov 9. J Cell Sci. 2016. PMID: 27831494
-
Cell adhesion in Drosophila: versatility of cadherin and integrin complexes during development.Curr Opin Cell Biol. 2012 Oct;24(5):702-12. doi: 10.1016/j.ceb.2012.07.006. Epub 2012 Aug 28. Curr Opin Cell Biol. 2012. PMID: 22938782 Free PMC article. Review.
-
Drosophila p120catenin plays a supporting role in cell adhesion but is not an essential adherens junction component.J Cell Biol. 2003 Feb 3;160(3):433-49. doi: 10.1083/jcb.200211083. Epub 2003 Jan 27. J Cell Biol. 2003. PMID: 12551951 Free PMC article.
-
Embryonal Fyn-associated substrate (EFS) and CASS4: The lesser-known CAS protein family members.Gene. 2015 Oct 1;570(1):25-35. doi: 10.1016/j.gene.2015.06.062. Epub 2015 Jun 26. Gene. 2015. PMID: 26119091 Free PMC article. Review.
Cited by
-
P130Cas/bcar1 mediates zebrafish caudal vein plexus angiogenesis.Sci Rep. 2020 Sep 24;10(1):15589. doi: 10.1038/s41598-020-71753-w. Sci Rep. 2020. PMID: 32973180 Free PMC article.
-
The role of the cilium in normal and abnormal cell cycles: emphasis on renal cystic pathologies.Cell Mol Life Sci. 2013 Jun;70(11):1849-74. doi: 10.1007/s00018-012-1052-z. Epub 2012 Jul 11. Cell Mol Life Sci. 2013. PMID: 22782110 Free PMC article. Review.
-
Disease-modifying genes and monogenic disorders: experience in cystic fibrosis.Appl Clin Genet. 2014 Jul 10;7:133-46. doi: 10.2147/TACG.S18675. eCollection 2014. Appl Clin Genet. 2014. PMID: 25053892 Free PMC article. Review.
-
Nedd9 restrains renal cystogenesis in Pkd1-/- mice.Proc Natl Acad Sci U S A. 2014 Sep 2;111(35):12859-64. doi: 10.1073/pnas.1405362111. Epub 2014 Aug 19. Proc Natl Acad Sci U S A. 2014. PMID: 25139996 Free PMC article.
-
Late-Onset Alzheimer's Disease Genes and the Potentially Implicated Pathways.Curr Genet Med Rep. 2014 Mar 22;2(2):85-101. doi: 10.1007/s40142-014-0034-x. eCollection 2014. Curr Genet Med Rep. 2014. PMID: 24829845 Free PMC article. Review.
References
-
- Defilippi P, Di Stefano P, Cabodi S. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol. 2006;16:257–263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

