Structural basis for certain naturally occurring bioflavonoids to function as reducing co-substrates of cyclooxygenase I and II
- PMID: 20808785
- PMCID: PMC2925883
- DOI: 10.1371/journal.pone.0012316
Structural basis for certain naturally occurring bioflavonoids to function as reducing co-substrates of cyclooxygenase I and II
Abstract
Background: Recent studies showed that some of the dietary bioflavonoids can strongly stimulate the catalytic activity of cyclooxygenase (COX) I and II in vitro and in vivo, presumably by facilitating enzyme re-activation. In this study, we sought to understand the structural basis of COX activation by these dietary compounds.
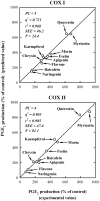
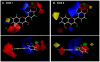
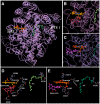
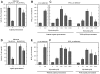
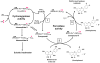
Methodology/principal findings: A combination of molecular modeling studies, biochemical analysis and site-directed mutagenesis assay was used as research tools. Three-dimensional quantitative structure-activity relationship analysis (QSAR/CoMFA) predicted that the ability of bioflavonoids to activate COX I and II depends heavily on their B-ring structure, a moiety known to be associated with strong antioxidant ability. Using the homology modeling and docking approaches, we identified the peroxidase active site of COX I and II as the binding site for bioflavonoids. Upon binding to this site, bioflavonoid can directly interact with hematin of the COX enzyme and facilitate the electron transfer from bioflavonoid to hematin. The docking results were verified by biochemical analysis, which reveals that when the cyclooxygenase activity of COXs is inhibited by covalent modification, myricetin can still stimulate the conversion of PGG(2) to PGE(2), a reaction selectively catalyzed by the peroxidase activity. Using the site-directed mutagenesis analysis, we confirmed that Q189 at the peroxidase site of COX II is essential for bioflavonoids to bind and re-activate its catalytic activity.
Conclusions/significance: These findings provide the structural basis for bioflavonoids to function as high-affinity reducing co-substrates of COXs through binding to the peroxidase active site, facilitating electron transfer and enzyme re-activation.
Conflict of interest statement
Figures

Similar articles
-
Inhibition of cyclooxygenase by blocking the reducing cosubstrate at the peroxidase site: Discovery of galangin as a novel cyclooxygenase inhibitor.Eur J Pharmacol. 2021 May 15;899:174036. doi: 10.1016/j.ejphar.2021.174036. Epub 2021 Mar 15. Eur J Pharmacol. 2021. PMID: 33737009
-
Strong activation of cyclooxygenase I and II catalytic activity by dietary bioflavonoids.J Lipid Res. 2008 Dec;49(12):2557-70. doi: 10.1194/jlr.M800358-JLR200. Epub 2008 Jul 26. J Lipid Res. 2008. PMID: 18660529 Free PMC article.
-
Usefulness of molecular modeling in characterizing the ligand-binding sites of proteins: experience with human PDI, PDIp and COX.Curr Med Chem. 2013;20(31):3840-54. doi: 10.2174/09298673113209990207. Curr Med Chem. 2013. PMID: 23931275 Review.
-
Isoxazole-Based-Scaffold Inhibitors Targeting Cyclooxygenases (COXs).ChemMedChem. 2016 Jun 6;11(11):1172-87. doi: 10.1002/cmdc.201500439. Epub 2016 May 2. ChemMedChem. 2016. PMID: 27136372
-
Computer aided drug design approaches to develop cyclooxygenase based novel anti-inflammatory and anti-cancer drugs.Curr Pharm Des. 2007;13(34):3505-17. doi: 10.2174/138161207782794275. Curr Pharm Des. 2007. PMID: 18220787 Review.
Cited by
-
Quantitative analysis of vasodilatory action of quercetin on intramural coronary resistance arteries of the rat in vitro.PLoS One. 2014 Aug 21;9(8):e105587. doi: 10.1371/journal.pone.0105587. eCollection 2014. PLoS One. 2014. PMID: 25144688 Free PMC article.
-
Impact of wines and wine constituents on cyclooxygenase-1, cyclooxygenase-2, and 5-lipoxygenase catalytic activity.Mediators Inflamm. 2014;2014:178931. doi: 10.1155/2014/178931. Epub 2014 May 29. Mediators Inflamm. 2014. PMID: 24976682 Free PMC article.
-
Rational Design of Novel Pyridinol-Fused Ring Acetaminophen Analogues.ACS Med Chem Lett. 2013 Aug 8;4(8):710-714. doi: 10.1021/ml4000904. ACS Med Chem Lett. 2013. PMID: 24482730 Free PMC article.
-
Mechanism for the reactivation of the peroxidase activity of human cyclooxygenases: investigation using phenol as a reducing cosubstrate.Sci Rep. 2020 Sep 16;10(1):15187. doi: 10.1038/s41598-020-71237-x. Sci Rep. 2020. PMID: 32938962 Free PMC article.
-
Stimulation of the Production of Prostaglandin E₂ by Ethyl Gallate, a Natural Phenolic Compound Richly Contained in Longan.Biomolecules. 2018 Sep 6;8(3):91. doi: 10.3390/biom8030091. Biomolecules. 2018. PMID: 30200641 Free PMC article.
References
-
- Hamberg M, Samuelsson B. Oxygenation of unsaturated fatty acids by the vesicular gland of sheep. J Biol Chem. 1967;242:5344–5354. - PubMed
-
- Miyamoto T, Ogino N, Yamamoto S, Hayaishi O. Purification of prostaglandin endoperoxide synthetase from bovine vesicular gland microsomes. J Biol Chem. 1976;251:2629–2636. - PubMed
-
- Kurumbail RG, Kiefer JR, Marnett LJ. Cyclooxygenase enzymes: catalysis and inhibition. Curr Opin Struct Biol. 2001;22:752–760. - PubMed
-
- Regan JW. EP2 and EP4 prostanoid receptor signaling. Life Sci. 2003;74:143–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

