Altered ultrasonic vocalization and impaired learning and memory in Angelman syndrome mouse model with a large maternal deletion from Ube3a to Gabrb3
- PMID: 20808828
- PMCID: PMC2924885
- DOI: 10.1371/journal.pone.0012278
Altered ultrasonic vocalization and impaired learning and memory in Angelman syndrome mouse model with a large maternal deletion from Ube3a to Gabrb3
Abstract
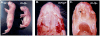
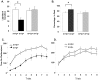
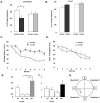
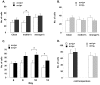
Angelman syndrome (AS) is a neurobehavioral disorder associated with mental retardation, absence of language development, characteristic electroencephalography (EEG) abnormalities and epilepsy, happy disposition, movement or balance disorders, and autistic behaviors. The molecular defects underlying AS are heterogeneous, including large maternal deletions of chromosome 15q11-q13 (70%), paternal uniparental disomy (UPD) of chromosome 15 (5%), imprinting mutations (rare), and mutations in the E6-AP ubiquitin ligase gene UBE3A (15%). Although patients with UBE3A mutations have a wide spectrum of neurological phenotypes, their features are usually milder than AS patients with deletions of 15q11-q13. Using a chromosomal engineering strategy, we generated mutant mice with a 1.6-Mb chromosomal deletion from Ube3a to Gabrb3, which inactivated the Ube3a and Gabrb3 genes and deleted the Atp10a gene. Homozygous deletion mutant mice died in the perinatal period due to a cleft palate resulting from the null mutation in Gabrb3 gene. Mice with a maternal deletion (m-/p+) were viable and did not have any obvious developmental defects. Expression analysis of the maternal and paternal deletion mice confirmed that the Ube3a gene is maternally expressed in brain, and showed that the Atp10a and Gabrb3 genes are biallelically expressed in all brain sub-regions studied. Maternal (m-/p+), but not paternal (m+/p-), deletion mice had increased spontaneous seizure activity and abnormal EEG. Extensive behavioral analyses revealed significant impairment in motor function, learning and memory tasks, and anxiety-related measures assayed in the light-dark box in maternal deletion but not paternal deletion mice. Ultrasonic vocalization (USV) recording in newborns revealed that maternal deletion pups emitted significantly more USVs than wild-type littermates. The increased USV in maternal deletion mice suggests abnormal signaling behavior between mothers and pups that may reflect abnormal communication behaviors in human AS patients. Thus, mutant mice with a maternal deletion from Ube3a to Gabrb3 provide an AS mouse model that is molecularly more similar to the contiguous gene deletion form of AS in humans than mice with Ube3a mutation alone. These mice will be valuable for future comparative studies to mice with maternal deficiency of Ube3a alone.
Conflict of interest statement
Figures



Similar articles
-
Angelman syndrome and severe infections in a patient with de novo 15q11.2-q13.1 deletion and maternally inherited 2q21.3 microdeletion.Gene. 2013 Jan 10;512(2):453-5. doi: 10.1016/j.gene.2012.10.061. Epub 2012 Nov 1. Gene. 2013. PMID: 23124039
-
Hypersociability in the Angelman syndrome mouse model.Exp Neurol. 2017 Jul;293:137-143. doi: 10.1016/j.expneurol.2017.04.002. Epub 2017 Apr 11. Exp Neurol. 2017. PMID: 28411125 Free PMC article.
-
Angelman syndrome - insights into a rare neurogenetic disorder.Nat Rev Neurol. 2016 Oct;12(10):584-93. doi: 10.1038/nrneurol.2016.133. Epub 2016 Sep 12. Nat Rev Neurol. 2016. PMID: 27615419 Review.
-
Mice lacking the beta3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome.J Neurosci. 1998 Oct 15;18(20):8505-14. doi: 10.1523/JNEUROSCI.18-20-08505.1998. J Neurosci. 1998. PMID: 9763493 Free PMC article.
-
Parental imprinting and Angelman syndrome.Adv Neurol. 1999;79:421-9. Adv Neurol. 1999. PMID: 10514831 Review.
Cited by
-
Clinical and Neurobiological Relevance of Current Animal Models of Autism Spectrum Disorders.Biomol Ther (Seoul). 2016 May 1;24(3):207-43. doi: 10.4062/biomolther.2016.061. Biomol Ther (Seoul). 2016. PMID: 27133257 Free PMC article. Review.
-
Seizure-like activity in a juvenile Angelman syndrome mouse model is attenuated by reducing Arc expression.Proc Natl Acad Sci U S A. 2015 Apr 21;112(16):5129-34. doi: 10.1073/pnas.1504809112. Epub 2015 Apr 6. Proc Natl Acad Sci U S A. 2015. PMID: 25848016 Free PMC article.
-
Synaptic plasticity in mouse models of autism spectrum disorders.Korean J Physiol Pharmacol. 2012 Dec;16(6):369-78. doi: 10.4196/kjpp.2012.16.6.369. Epub 2012 Dec 10. Korean J Physiol Pharmacol. 2012. PMID: 23269898 Free PMC article.
-
Linoleic acid improves PIEZO2 dysfunction in a mouse model of Angelman Syndrome.Nat Commun. 2023 Mar 1;14(1):1167. doi: 10.1038/s41467-023-36818-0. Nat Commun. 2023. PMID: 36859399 Free PMC article.
-
Medial Preoptic Area Modulates Courtship Ultrasonic Vocalization in Adult Male Mice.Neurosci Bull. 2019 Aug;35(4):697-708. doi: 10.1007/s12264-019-00365-w. Epub 2019 Mar 21. Neurosci Bull. 2019. PMID: 30900143 Free PMC article.
References
-
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. - PubMed
-
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. - PubMed
-
- Veenstra-Vanderweele J, Christian SL, Cook EH., Jr Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. - PubMed
-
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

