Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos
- PMID: 20808841
- PMCID: PMC2924899
- DOI: 10.1371/journal.pone.0012301
Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos
Abstract
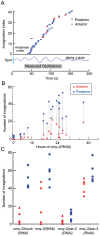
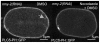
Asymmetric positioning of the mitotic spindle in C. elegans embryos is mediated by force-generating complexes that are anchored at the plasma membrane and that pull on microtubules growing out from the spindle poles. Although asymmetric distribution of the force generators is thought to underlie asymmetric positioning of the spindle, the number and location of the force generators has not been well defined. In particular, it has not been possible to visualize individual force generating events at the cortex. We discovered that perturbation of the acto-myosin cortex leads to the formation of long membrane invaginations that are pulled from the plasma membrane toward the spindle poles. Several lines of evidence show that the invaginations, which also occur in unperturbed embryos though at lower frequency, are pulled by the same force generators responsible for spindle positioning. Thus, the invaginations serve as a tool to localize the sites of force generation at the cortex and allow us to estimate a lower limit on the number of cortical force generators within the cell.
Conflict of interest statement
Figures


Similar articles
-
Clathrin regulates centrosome positioning by promoting acto-myosin cortical tension in C. elegans embryos.Development. 2014 Jul;141(13):2712-23. doi: 10.1242/dev.107508. Development. 2014. PMID: 24961801
-
Asymmetric division: motor persistence pays off.Curr Biol. 2006 Dec 19;16(24):R1021-3. doi: 10.1016/j.cub.2006.11.007. Curr Biol. 2006. PMID: 17174905
-
Modeling microtubule-mediated forces and centrosome positioning in Caenorhabditis elegans embryos.Methods Cell Biol. 2010;97:437-53. doi: 10.1016/S0091-679X(10)97023-4. Methods Cell Biol. 2010. PMID: 20719284
-
Mechanisms of spindle positioning: cortical force generators in the limelight.Curr Opin Cell Biol. 2013 Dec;25(6):741-8. doi: 10.1016/j.ceb.2013.07.008. Epub 2013 Aug 16. Curr Opin Cell Biol. 2013. PMID: 23958212 Review.
-
[Mechanisms of cell division: lessons from a nematode].Med Sci (Paris). 2003 Jun-Jul;19(6-7):735-42. doi: 10.1051/medsci/20031967735. Med Sci (Paris). 2003. PMID: 12942445 Review. French.
Cited by
-
Dynein localization and pronuclear movement in the C. elegans zygote.Cytoskeleton (Hoboken). 2022 Dec;79(12):133-143. doi: 10.1002/cm.21733. Epub 2022 Oct 28. Cytoskeleton (Hoboken). 2022. PMID: 36214774 Free PMC article.
-
F-actin asymmetry and the endoplasmic reticulum-associated TCC-1 protein contribute to stereotypic spindle movements in the Caenorhabditis elegans embryo.Mol Biol Cell. 2013 Jul;24(14):2201-15. doi: 10.1091/mbc.E13-02-0076. Epub 2013 May 22. Mol Biol Cell. 2013. PMID: 23699393 Free PMC article.
-
Astral microtubules control redistribution of dynein at the cell cortex to facilitate spindle positioning.Cell Cycle. 2014;13(7):1162-70. doi: 10.4161/cc.28031. Epub 2014 Feb 10. Cell Cycle. 2014. PMID: 24553118 Free PMC article.
-
Epithelial polarity and spindle orientation: intersecting pathways.Philos Trans R Soc Lond B Biol Sci. 2013 Sep 23;368(1629):20130291. doi: 10.1098/rstb.2013.0291. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24062590 Free PMC article. Review.
-
Two populations of cytoplasmic dynein contribute to spindle positioning in C. elegans embryos.J Cell Biol. 2017 Sep 4;216(9):2777-2793. doi: 10.1083/jcb.201607038. Epub 2017 Jul 24. J Cell Biol. 2017. PMID: 28739679 Free PMC article.
References
-
- Dogterom M, Kerssemakers JW, Romet-Lemonne G, Janson ME. Force generation by dynamic microtubules. Curr Opin Cell Biol. 2005;17:67–74. - PubMed
-
- Kunda P, Pelling AE, Liu T, Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18:91–101. - PubMed
-
- Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Current Biology. 2004;14:1181–1186. - PubMed
-
- Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

