Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells
- PMID: 20808852
- PMCID: PMC2924374
- DOI: 10.1371/journal.ppat.1001063
Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells
Abstract
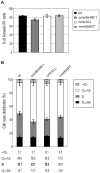
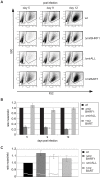
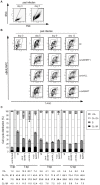
Cellular and viral microRNAs (miRNAs) are involved in many different processes of key importance and more than 10,000 miRNAs have been identified so far. In general, relatively little is known about their biological functions in mammalian cells because their phenotypic effects are often mild and many of their targets still await identification. The recent discovery that Epstein-Barr virus (EBV) and other herpesviruses produce their own, barely conserved sets of miRNAs suggests that these viruses usurp the host RNA silencing machinery to their advantage in contrast to the antiviral roles of RNA silencing in plants and insects. We have systematically introduced mutations in EBV's precursor miRNA transcripts to prevent their subsequent processing into mature viral miRNAs. Phenotypic analyses of these mutant derivatives of EBV revealed that the viral miRNAs of the BHRF1 locus inhibit apoptosis and favor cell cycle progression and proliferation during the early phase of infected human primary B cells. Our findings also indicate that EBV's miRNAs are not needed to control the exit from latency. The phenotypes of viral miRNAs uncovered by this genetic analysis indicate that they contribute to EBV-associated cellular transformation rather than regulate viral genes of EBV's lytic phase.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus.PLoS Pathog. 2011 Feb;7(2):e1001294. doi: 10.1371/journal.ppat.1001294. Epub 2011 Feb 17. PLoS Pathog. 2011. PMID: 21379335 Free PMC article.
-
Multiple Viral microRNAs Regulate Interferon Release and Signaling Early during Infection with Epstein-Barr Virus.mBio. 2021 Mar 30;12(2):e03440-20. doi: 10.1128/mBio.03440-20. mBio. 2021. PMID: 33785626 Free PMC article.
-
First Days in the Life of Naive Human B Lymphocytes Infected with Epstein-Barr Virus.mBio. 2019 Sep 17;10(5):e01723-19. doi: 10.1128/mBio.01723-19. mBio. 2019. PMID: 31530670 Free PMC article.
-
Human B cells on their route to latent infection--early but transient expression of lytic genes of Epstein-Barr virus.Eur J Cell Biol. 2012 Jan;91(1):65-9. doi: 10.1016/j.ejcb.2011.01.014. Epub 2011 Mar 29. Eur J Cell Biol. 2012. PMID: 21450364 Review.
-
Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases.Front Immunol. 2020 Mar 3;11:367. doi: 10.3389/fimmu.2020.00367. eCollection 2020. Front Immunol. 2020. PMID: 32194570 Free PMC article. Review.
Cited by
-
Viral miRNA regulation of host gene expression.Semin Cell Dev Biol. 2023 Sep 15;146:2-19. doi: 10.1016/j.semcdb.2022.11.007. Epub 2022 Nov 30. Semin Cell Dev Biol. 2023. PMID: 36463091 Free PMC article. Review.
-
Latent Epstein-Barr virus can inhibit apoptosis in B cells by blocking the induction of NOXA expression.PLoS One. 2011;6(12):e28506. doi: 10.1371/journal.pone.0028506. Epub 2011 Dec 9. PLoS One. 2011. PMID: 22174825 Free PMC article.
-
Epstein-Barr virus latent genes.Exp Mol Med. 2015 Jan 23;47(1):e131. doi: 10.1038/emm.2014.84. Exp Mol Med. 2015. PMID: 25613728 Free PMC article. Review.
-
Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing.J Exp Med. 2016 Sep 19;213(10):2065-80. doi: 10.1084/jem.20160248. Epub 2016 Sep 12. J Exp Med. 2016. PMID: 27621419 Free PMC article.
-
MicroRNAs are minor constituents of extracellular vesicles that are rarely delivered to target cells.PLoS Genet. 2021 Dec 6;17(12):e1009951. doi: 10.1371/journal.pgen.1009951. eCollection 2021 Dec. PLoS Genet. 2021. PMID: 34871319 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

