Small molecule inhibitors of DNA repair nuclease activities of APE1
- PMID: 20809131
- PMCID: PMC2956791
- DOI: 10.1007/s00018-010-0488-2
Small molecule inhibitors of DNA repair nuclease activities of APE1
Abstract
APE1 is a multifunctional protein that possesses several nuclease activities, including the ability to incise at apurinic/apyrimidinic (AP) sites in DNA or RNA, to excise 3'-blocking termini from DNA ends, and to cleave at certain oxidized base lesions in DNA. Pre-clinical and clinical data indicate a role for APE1 in the pathogenesis of cancer and in resistance to DNA-interactive drugs, particularly monofunctional alkylators and antimetabolites. In an effort to improve the efficacy of therapeutic compounds, such as temozolomide, groups have begun to develop high-throughput screening assays and to identify small molecule inhibitors against APE1 repair nuclease activities. It is envisioned that such inhibitors will be used in combinatorial treatment paradigms to enhance the efficacy of DNA-interactive drugs that introduce relevant cytotoxic DNA lesions. In this review, we summarize the current state of the efforts to design potent and selective inhibitors against APE1 AP site incision activity.
Figures



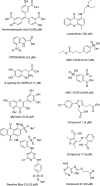
Similar articles
-
Inhibitors of nuclease and redox activity of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1).Bioorg Med Chem. 2017 May 1;25(9):2531-2544. doi: 10.1016/j.bmc.2017.01.028. Epub 2017 Jan 21. Bioorg Med Chem. 2017. PMID: 28161249 Review.
-
Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors.J Med Chem. 2009 Jan 8;52(1):20-32. doi: 10.1021/jm800739m. J Med Chem. 2009. PMID: 19072053
-
Isolation of a small molecule inhibitor of DNA base excision repair.Nucleic Acids Res. 2005 Aug 19;33(15):4711-24. doi: 10.1093/nar/gki781. Print 2005. Nucleic Acids Res. 2005. PMID: 16113242 Free PMC article.
-
Small-molecule inhibitors of APE1 DNA repair function: an overview.Curr Mol Pharmacol. 2012 Jan;5(1):14-35. Curr Mol Pharmacol. 2012. PMID: 22122462 Review.
-
Diverse small molecule inhibitors of human apurinic/apyrimidinic endonuclease APE1 identified from a screen of a large public collection.PLoS One. 2012;7(10):e47974. doi: 10.1371/journal.pone.0047974. Epub 2012 Oct 23. PLoS One. 2012. PMID: 23110144 Free PMC article.
Cited by
-
Visualization and Comparison of the Level of Apurinic/Apyrimidinic Endonuclease 1 in Live Normal/Cancerous and Neuron Cells with a Fluorescent Nanoprobe.Molecules. 2023 May 7;28(9):3935. doi: 10.3390/molecules28093935. Molecules. 2023. PMID: 37175345 Free PMC article.
-
An APE1 inhibitor reveals critical roles of the redox function of APE1 in KSHV replication and pathogenic phenotypes.PLoS Pathog. 2017 Apr 5;13(4):e1006289. doi: 10.1371/journal.ppat.1006289. eCollection 2017 Apr. PLoS Pathog. 2017. PMID: 28380040 Free PMC article.
-
Base excision repair: contribution to tumorigenesis and target in anticancer treatment paradigms.Curr Med Chem. 2012;19(23):3922-36. doi: 10.2174/092986712802002581. Curr Med Chem. 2012. PMID: 22788768 Free PMC article. Review.
-
Revisiting Two Decades of Research Focused on Targeting APE1 for Cancer Therapy: The Pros and Cons.Cells. 2023 Jul 20;12(14):1895. doi: 10.3390/cells12141895. Cells. 2023. PMID: 37508559 Free PMC article. Review.
-
The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once.DNA Repair (Amst). 2014 Jul;19:95-107. doi: 10.1016/j.dnarep.2014.03.022. Epub 2014 Apr 19. DNA Repair (Amst). 2014. PMID: 24754999 Free PMC article. Review.
References
-
- Friedberg EC. A brief history of the DNA repair field. Cell Res. 2008;18:3–7. - PubMed
-
- Dulbecco R. Reactivation of ultra-violet-inactivated bacteriophage by visible light. Nature. 1949;163:949. - PubMed
-
- Rupert CS. Enzymatic photoreactivation: overview. Basic Life Sci. 1975;5A:73–87. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

