A continuous mapping of sleep states through association of EEG with a mesoscale cortical model
- PMID: 20809258
- PMCID: PMC3058368
- DOI: 10.1007/s10827-010-0272-1
A continuous mapping of sleep states through association of EEG with a mesoscale cortical model
Abstract
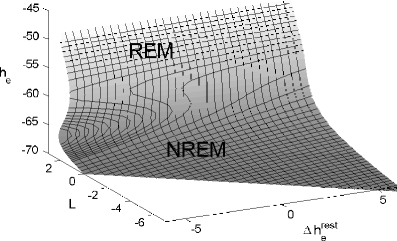
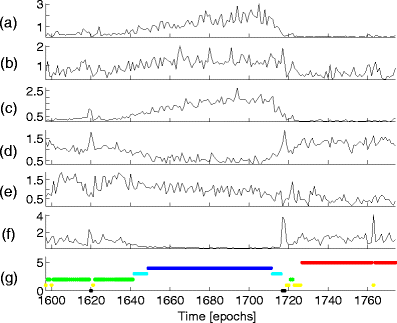
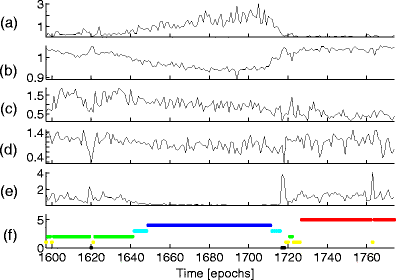
Here we show that a mathematical model of the human sleep cycle can be used to obtain a detailed description of electroencephalogram (EEG) sleep stages, and we discuss how this analysis may aid in the prediction and prevention of seizures during sleep. The association between EEG data and the cortical model is found via locally linear embedding (LLE), a method of dimensionality reduction. We first show that LLE can distinguish between traditional sleep stages when applied to EEG data. It reliably separates REM and non-REM sleep and maps the EEG data to a low-dimensional output space where the sleep state changes smoothly over time. We also incorporate the concept of strongly connected components and use this as a method of automatic outlier rejection for EEG data. Then, by using LLE on a hybrid data set containing both sleep EEG and signals generated from the mesoscale cortical model, we quantify the relationship between the data and the mathematical model. This enables us to take any sample of sleep EEG data and associate it with a position among the continuous range of sleep states provided by the model; we can thus infer a trajectory of states as the subject sleeps. Lastly, we show that this method gives consistent results for various subjects over a full night of sleep and can be done in real time.
Figures








References
-
- Ataee, P., Yazdani, A., Setarehdan, S. K., & Noubari, H. A. (2007). Manifold learning applied on EEG signal of the epileptic patients for detection of normal and pre-seizure states. In Proceedings of the 29th Annual International Conference of the IEEE EMBS (pp. 5489–5492). - PubMed
-
- Bojak, I., & Liley, D. T. (2005). Modeling the effects of anesthesia on the electroencephalogram. Physical Review E, 71(041902). - PubMed
-
- Corsi-Cabrera M, Guevara MA, Río-Portilla YD, Arce C, Villanueva-Hernández Y. EEG bands during wakefulness, slow-wave and paradoxical sleep as a result of principal component analysis in man. SLEEP. 2000;23(6):1–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

