Combining SELEX screening and rational design to develop light-up fluorophore-RNA aptamer pairs for RNA tagging
- PMID: 20809562
- PMCID: PMC3044212
- DOI: 10.1021/cb1001894
Combining SELEX screening and rational design to develop light-up fluorophore-RNA aptamer pairs for RNA tagging
Abstract
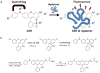
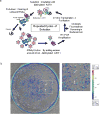
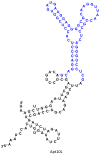
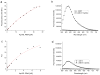
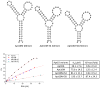
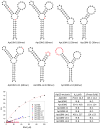
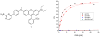
We report here a new small molecule fluorogen and RNA aptamer pair for RNA labeling. The small-molecule fluorogen is designed on the basis of fluorescently quenched sulforhodamine dye. The SELEX (Systematic Evolution of Ligands by EXponential enrichment) procedure and fluorescence screening in E. coli have been applied to discover the aptamer that can specifically activate the fluorogen with micromolar binding affinity. The systematic mutation and truncation study on the aptamer structure determined the minimum binding domain of the aptamer. A series of rationally modified fluorogen analogues have been made to probe the interacting groups of fluorogen with the aptamer. These results led to the design of a much improved fluorogen ASR 7 that displayed a 33-fold increase in the binding affinity for the selected aptamer in comparison to the original ASR 1 and an 88-fold increase in the fluorescence emission after the aptamer binding. This study demonstrates the value of combining in vitro SELEX and E. coli fluorescence screening with rational modifications in discovering and optimizing new fluorogen-RNA aptamer labeling pairs.
Figures

References
-
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. - PubMed
-
- Marks KM, Nolan GP. Chemical labeling strategies for cell biology. Nat Methods. 2006;3:591–596. - PubMed
-
- Johnsson N, Johnsson K. Chemical tools for biomolecular imaging. ACS Chem Biol. 2007;2:31–38. - PubMed
-
- Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. - PubMed
-
- Dragulescu-Andrasi A, Rao J. Chemical labeling of protein in living cells. ChemBioChem. 2007;8:1099–1101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

