Open access high throughput drug discovery in the public domain: a Mount Everest in the making
- PMID: 20809896
- PMCID: PMC3716285
- DOI: 10.2174/138920110792927757
Open access high throughput drug discovery in the public domain: a Mount Everest in the making
Abstract
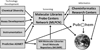
High throughput screening (HTS) facilitates screening large numbers of compounds against a biochemical target of interest using validated biological or biophysical assays. In recent years, a significant number of drugs in clinical trails originated from HTS campaigns, validating HTS as a bona fide mechanism for hit finding. In the current drug discovery landscape, the pharmaceutical industry is embracing open innovation strategies with academia to maximize their research capabilities and to feed their drug discovery pipeline. The goals of academic research have therefore expanded from target identification and validation to probe discovery, chemical genomics, and compound library screening. This trend is reflected in the emergence of HTS centers in the public domain over the past decade, ranging in size from modestly equipped academic screening centers to well endowed Molecular Libraries Probe Centers Network (MLPCN) centers funded by the NIH Roadmap initiative. These centers facilitate a comprehensive approach to probe discovery in academia and utilize both classical and cutting-edge assay technologies for executing primary and secondary screening campaigns. The various facets of academic HTS centers as well as their implications on technology transfer and drug discovery are discussed, and a roadmap for successful drug discovery in the public domain is presented. New lead discovery against therapeutic targets, especially those involving the rare and neglected diseases, is indeed a Mount Everestonian size task, and requires diligent implementation of pharmaceutical industry's best practices for a successful outcome.
Figures



References
-
- Munos B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009;8(12):959–968. - PubMed
-
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5(12):993–996. - PubMed
-
- Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1(9):727–730. - PubMed
-
- Sakharkar MK, Sakharkar KR. Targetability of human disease genes. Curr. Drug Discov. Technol. 2007;4(1):48–58. - PubMed
-
- Knowles J, Gromo G. A guide to drug discovery: Target selection in drug discovery. Nat. Rev. Drug Discov. 2003;2(1):63–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

