Meis1 specifies positional information in the retina and tectum to organize the zebrafish visual system
- PMID: 20809932
- PMCID: PMC2939508
- DOI: 10.1186/1749-8104-5-22
Meis1 specifies positional information in the retina and tectum to organize the zebrafish visual system
Abstract
Background: During visual system development, multiple signalling pathways cooperate to specify axial polarity within the retina and optic tectum. This information is required for the topographic mapping of retinal ganglion cell axons on the tectum. Meis1 is a TALE-class homeodomain transcription factor known to specify anterior-posterior identity in the hindbrain, but its role in visual system patterning has not been investigated.
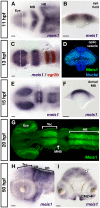
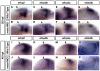
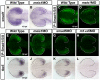
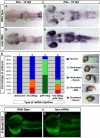
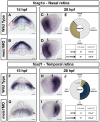
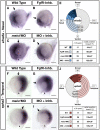
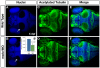
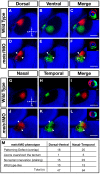
Results: meis1 is expressed in both the presumptive retina and tectum. An analysis of retinal patterning reveals that Meis1 is required to correctly specify both dorsal-ventral and nasal-temporal identity in the zebrafish retina. Meis1-knockdown results in a loss of smad1 expression and an upregulation in follistatin expression, thereby causing lower levels of Bmp signalling and a partial ventralization of the retina. Additionally, Meis1-deficient embryos exhibit ectopic Fgf signalling in the developing retina and a corresponding loss of temporal identity. Meis1 also positively regulates ephrin gene expression in the tectum. Consistent with these patterning phenotypes, a knockdown of Meis1 ultimately results in retinotectal mapping defects.
Conclusions: In this work we describe a novel role for Meis1 in regulating Bmp signalling and in specifying temporal identity in the retina. By patterning both the retina and tectum, Meis1 plays an important role in establishing the retinotectal map and organizing the visual system.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

