Overlap and effective size of the human CD8+ T cell receptor repertoire
- PMID: 20811043
- PMCID: PMC3212437
- DOI: 10.1126/scitranslmed.3001442
Overlap and effective size of the human CD8+ T cell receptor repertoire
Abstract
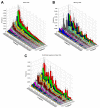
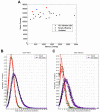
Diversity in T lymphocyte antigen receptors is generated by somatic rearrangement of T cell receptor (TCR) genes and is concentrated within the third complementarity-determining region 3 (CDR3) of each chain of the TCR heterodimer. We sequenced the CDR3 regions from millions of rearranged TCR beta chain genes in naïve and memory CD8(+) T cells of seven adults. The CDR3 sequence repertoire realized in each individual is strongly biased toward specific V(beta)-J(beta) pair utilization, dominated by sequences containing few inserted nucleotides, and drawn from a defined subset comprising less than 0.1% of the estimated 5 x 10(11) possible sequences. Surprisingly, the overlap in the naïve CD8(+) CDR3 sequence repertoires of any two of the individuals is approximately 7000-fold larger than predicted and appears to be independent of the degree of human leukocyte antigen matching.
Figures


References
-
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988 Aug 4;334(6181):395–402. - PubMed
-
- Monod M Yousfi, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004 Aug 4;20(Suppl 1):i379–385. - PubMed
-
- Malissen M, McCoy C, Blanc D, et al. Direct evidence for chromosomal inversion during T-cell receptor beta-gene rearrangements. Nature. 1986 Jan 2-8;319(6048):28–33. - PubMed
-
- Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008 Mar;8(3):231–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

