Evaluating genome-scale approaches to eukaryotic DNA replication
- PMID: 20811343
- PMCID: PMC2962615
- DOI: 10.1038/nrg2830
Evaluating genome-scale approaches to eukaryotic DNA replication
Abstract
Mechanisms regulating where and when eukaryotic DNA replication initiates remain a mystery. Recently, genome-scale methods have been brought to bear on this problem. The identification of replication origins and their associated proteins in yeasts is a well-integrated investigative tool, but corresponding data sets from multicellular organisms are scarce. By contrast, standardized protocols for evaluating replication timing have generated informative data sets for most eukaryotic systems. Here, I summarize the genome-scale methods that are most frequently used to analyse replication in eukaryotes, the kinds of questions each method can address and the technical hurdles that must be overcome to gain a complete understanding of the nature of eukaryotic replication origins.
Figures
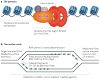
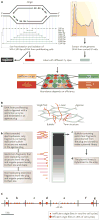
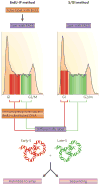
References
-
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

