Bacterial charity work leads to population-wide resistance
- PMID: 20811456
- PMCID: PMC2936489
- DOI: 10.1038/nature09354
Bacterial charity work leads to population-wide resistance
Abstract
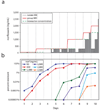
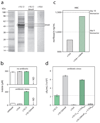
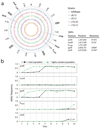
Bacteria show remarkable adaptability in the face of antibiotic therapeutics. Resistance alleles in drug target-specific sites and general stress responses have been identified in individual end-point isolates. Less is known, however, about the population dynamics during the development of antibiotic-resistant strains. Here we follow a continuous culture of Escherichia coli facing increasing levels of antibiotic and show that the vast majority of isolates are less resistant than the population as a whole. We find that the few highly resistant mutants improve the survival of the population's less resistant constituents, in part by producing indole, a signalling molecule generated by actively growing, unstressed cells. We show, through transcriptional profiling, that indole serves to turn on drug efflux pumps and oxidative-stress protective mechanisms. The indole production comes at a fitness cost to the highly resistant isolates, and whole-genome sequencing reveals that this bacterial altruism is made possible by drug-resistance mutations unrelated to indole production. This work establishes a population-based resistance mechanism constituting a form of kin selection whereby a small number of resistant mutants can, at some cost to themselves, provide protection to other, more vulnerable, cells, enhancing the survival capacity of the overall population in stressful environments.
Figures

Comment in
-
Microbiology: Altruistic defence.Nature. 2010 Sep 2;467(7311):34-5. doi: 10.1038/467034a. Nature. 2010. PMID: 20811443 No abstract available.
References
-
- Albert TJ, et al. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2(12):951–953. - PubMed
-
- Bjedov I, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300(5624):1404–1409. - PubMed
-
- Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

