Mutational characterization of the bile acid receptor TGR5 in primary sclerosing cholangitis
- PMID: 20811628
- PMCID: PMC2928275
- DOI: 10.1371/journal.pone.0012403
Mutational characterization of the bile acid receptor TGR5 in primary sclerosing cholangitis
Abstract
Background: TGR5, the G protein-coupled bile acid receptor 1 (GPBAR1), has been linked to inflammatory pathways as well as bile homeostasis, and could therefore be involved in primary sclerosing cholangitis (PSC) a chronic inflammatory bile duct disease. We aimed to extensively investigate TGR5 sequence variation in PSC, as well as functionally characterize detected variants.
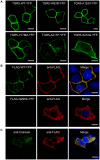
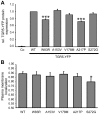
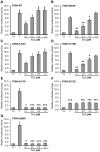
Methodology/principal findings: Complete resequencing of TGR5 was performed in 267 PSC patients and 274 healthy controls. Six nonsynonymous mutations were identified in addition to 16 other novel single-nucleotide polymorphisms. To investigate the impact from the nonsynonymous variants on TGR5, we created a receptor model, and introduced mutated TGR5 constructs into human epithelial cell lines. By using confocal microscopy, flow cytometry and a cAMP-sensitive luciferase assay, five of the nonsynonymous mutations (W83R, V178M, A217P, S272G and Q296X) were found to reduce or abolish TGR5 function. Fine-mapping of the previously reported PSC and UC associated locus at chromosome 2q35 in large patient panels revealed an overall association between the TGR5 single-nucleotide polymorphism rs11554825 and PSC (odds ratio = 1.14, 95% confidence interval: 1.03-1.26, p = 0.010) and UC (odds ratio = 1.19, 95% confidence interval 1.11-1.27, p = 8.5 x 10(-7)), but strong linkage disequilibrium precluded demarcation of TGR5 from neighboring genes.
Conclusions/significance: Resequencing of TGR5 along with functional investigations of novel variants provided unique insight into an important candidate gene for several inflammatory and metabolic conditions. While significant TGR5 associations were detected in both UC and PSC, further studies are needed to conclusively define the role of TGR5 variation in these diseases.
Conflict of interest statement
Figures



References
-
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun. 2002;298:714–719. - PubMed
-
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. - PubMed
-
- Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. - PubMed
-
- Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, et al. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191:197–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

