The RhoA GEF Syx is a target of Rnd3 and regulated via a Raf1-like ubiquitin-related domain
- PMID: 20811643
- PMCID: PMC2928299
- DOI: 10.1371/journal.pone.0012409
The RhoA GEF Syx is a target of Rnd3 and regulated via a Raf1-like ubiquitin-related domain
Abstract
Background: Rnd3 (RhoE) protein belongs to the unique branch of Rho family GTPases that has low intrinsic GTPase activity and consequently remains constitutively active [1], [2]. The current consensus is that Rnd1 and Rnd3 function as important antagonists of RhoA signaling primarily by activating the ubiquitous p190 RhoGAP [3], but not by inhibiting the ROCK family kinases.
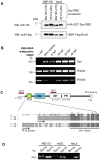
Methodology/principal findings: Rnd3 is abundant in mouse embryonic stem (mES) cells and in an unbiased two-step affinity purification screen we identified a new Rnd3 target, termed synectin-binding RhoA exchange factor (Syx), by mass spectrometry. The Syx interaction with Rnd3 does not occur through the Syx DH domain but utilizes a region similar to the classic Raf1 Ras-binding domain (RBD), and most closely related to those in RGS12 and RGS14. We show that Syx behaves as a genuine effector of Rnd3 (and perhaps Rnd1), with binding characteristics similar to p190-RhoGAP. Morpholino-oligonucleotide knockdown of Syx in zebrafish at the one cell stage resulted in embryos with shortened anterior-posterior body axis: this phenotype was effectively rescued by introducing mouse Syx1b mRNA. A Rnd3-binding defective mutant of Syx1b mutated in the RBD (E164A/R165D) was more potent in rescuing the embryonic defects than wild-type Syx1b, showing that Rnd3 negatively regulates Syx activity in vivo.
Conclusions/significance: This study uncovers a well defined Rnd3 effector Syx which is widely expressed and directly impacts RhoA activation. Experiments conducted in vivo indicate that Rnd3 negatively regulates Syx, and that as a RhoA-GEF it plays a key role in early embryonic cell shape changes. Thus a connection to signaling via the planar cell polarity pathway is suggested.
Conflict of interest statement
Figures





References
-
- Chardin P. Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol. 2006;7:54–62. - PubMed
-
- Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–842. - PubMed
-
- Stam JC, Collard JG. The DH protein family, exchange factors for Rho-like GTPases. Prog Mol Subcell Biol. 1999;22:51–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

