Cellular arrays for large-scale analysis of transcription factor activity
- PMID: 20812256
- PMCID: PMC3022829
- DOI: 10.1002/bit.22916
Cellular arrays for large-scale analysis of transcription factor activity
Abstract
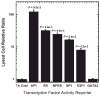
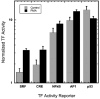
Identifying molecular mechanisms or therapeutic targets is typically based on large-scale cellular analysis that measures the abundance of mRNA or protein; however, abundance does not necessarily correlate with activity. We report a method for direct large-scale quantification of active pathways that employs a cellular array with parallel gene delivery of constructs that report pathway activity. The reporter constructs encode luciferase, whose expression is influenced by binding of transcription factors (TFs), which are the downstream targets of signaling pathways. Luciferase levels are quantified by bioluminescence imaging (BLI), which allows for rapid, non-invasive measurements. Activity profiles by BLI of 32 TFs were robust, consistent, and reproducible, and correlated with standard cell lysis techniques. The array identified five TFs with differential activity during phorbol-12-myristate-13-acetate (PMA)-induced differentiation of breast cancer cells. A system for rapid, large-scale, BLI quantification of pathway activity provides an enabling technology for mechanistic studies of cellular responses and processes.
2010 Wiley Periodicals, Inc.
Figures



References
-
- Abdel-Mageed AB, Agrawal KC. Activation of nuclear factor kappaB: Potential role in metallothionein-mediated mitogenic response. Cancer Res. 1998;58(11):2335–2338. - PubMed
-
- Adiseshaiah P, Peddakama S, Zhang Q, Kalvakolanu DV, Reddy SP. Mitogen regulated induction of FRA-1 proto-oncogene is controlled by the transcription factors binding to both serum and TPA response elements. Oncogene. 2005;24(26):4193–4205. - PubMed
-
- Ariazi EA, Kraus RJ, Farrell ML, Jordan VC, Mertz JE. Estrogenrelated receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol Cancer Res. 2007;5(1):71–85. - PubMed
-
- Bale TL, Dorsa DM. NGF, cyclic AMP, and phorbol esters regulate oxytocin receptor gene transcription in SK-N-SH and MCF7 cells. Brain Res Mol Brain Res. 1998;53(1–2):130–137. - PubMed
-
- Barnes PJ. Transcription factors in airway diseases. Lab Invest. 2006;86(9):867–872. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

