Distinct patterns of mitochondrial genome diversity in bonobos (Pan paniscus) and humans
- PMID: 20813043
- PMCID: PMC2942848
- DOI: 10.1186/1471-2148-10-270
Distinct patterns of mitochondrial genome diversity in bonobos (Pan paniscus) and humans
Abstract
Background: We have analyzed the complete mitochondrial genomes of 22 Pan paniscus (bonobo, pygmy chimpanzee) individuals to assess the detailed mitochondrial DNA (mtDNA) phylogeny of this close relative of Homo sapiens.
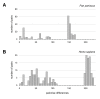
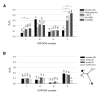
Results: We identified three major clades among bonobos that separated approximately 540,000 years ago, as suggested by Bayesian analysis. Incidentally, we discovered that the current reference sequence for bonobo likely is a hybrid of the mitochondrial genomes of two distant individuals. When comparing spectra of polymorphic mtDNA sites in bonobos and humans, we observed two major differences: (i) Of all 31 bonobo mtDNA homoplasies, i.e. nucleotide changes that occurred independently on separate branches of the phylogenetic tree, 13 were not homoplasic in humans. This indicates that at least a part of the unstable sites of the mitochondrial genome is species-specific and difficult to be explained on the basis of a mutational hotspot concept. (ii) A comparison of the ratios of non-synonymous to synonymous changes (dN/dS) among polymorphic positions in bonobos and in 4902 Homo sapiens mitochondrial genomes revealed a remarkable difference in the strength of purifying selection in the mitochondrial genes of the F0F1-ATPase complex. While in bonobos this complex showed a similar low value as complexes I and IV, human haplogroups displayed 2.2 to 7.6 times increased dN/dS ratios when compared to bonobos.
Conclusions: Some variants of mitochondrially encoded subunits of the ATPase complex in humans very likely decrease the efficiency of energy conversion leading to production of extra heat. Thus, we hypothesize that the species-specific release of evolutionary constraints for the mitochondrial genes of the proton-translocating ATPase is a consequence of altered heat homeostasis in modern humans.
Figures

References
-
- Xu X, Arnason U. A complete sequence of the mitochondrial genome of the western lowland gorilla. Mol Biol Evol. 1996;13:691–698. - PubMed
-
- Flynn SM, Carr SM. Interspecies hybridization on DNA resequencing microarrays: efficiency of sequence recovery and accuracy of SNP detection in human, ape, and codfish mitochondrial DNA genomes sequenced on a human-specific MitoChip. BMC Genomics. 2007;8:339. doi: 10.1186/1471-2164-8-339. - DOI - PMC - PubMed
-
- Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, Prüfer K, Siebauer M, Burbano HA, Ronan M, Rothberg JM, Egholm M, Rudan P, Brajković D, Kućan Z, Gusić I, Wikström M, Laakkonen L, Kelso J, Slatkin M, Pääbo S. A complete Neandertal mitochondrial genome sequence determined by highthroughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. - DOI - PMC - PubMed
-
- Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Schmitz R, Doronichev VB, Golovanova LV, de la Rasilla M, Fortea J, Rosas A, Pääbo S. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

