Identification of tumor-associated cassette exons in human cancer through EST-based computational prediction and experimental validation
- PMID: 20813049
- PMCID: PMC2941758
- DOI: 10.1186/1476-4598-9-230
Identification of tumor-associated cassette exons in human cancer through EST-based computational prediction and experimental validation
Abstract
Background: Many evidences report that alternative splicing, the mechanism which produces mRNAs and proteins with different structures and functions from the same gene, is altered in cancer cells. Thus, the identification and characterization of cancer-specific splice variants may give large impulse to the discovery of novel diagnostic and prognostic tumour biomarkers, as well as of new targets for more selective and effective therapies.
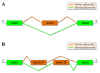
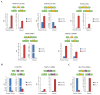
Results: We present here a genome-wide analysis of the alternative splicing pattern of human genes through a computational analysis of normal and cancer-specific ESTs from seventeen anatomical groups, using data available in AspicDB, a database resource for the analysis of alternative splicing in human. By using a statistical methodology, normal and cancer-specific genes, splice sites and cassette exons were predicted in silico. The condition association of some of the novel normal/tumoral cassette exons was experimentally verified by RT-qPCR assays in the same anatomical system where they were predicted. Remarkably, the presence in vivo of the predicted alternative transcripts, specific for the nervous system, was confirmed in patients affected by glioblastoma.
Conclusion: This study presents a novel computational methodology for the identification of tumor-associated transcript variants to be used as cancer molecular biomarkers, provides its experimental validation, and reports specific biomarkers for glioblastoma.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

