Spiroindolones, a potent compound class for the treatment of malaria
- PMID: 20813948
- PMCID: PMC3050001
- DOI: 10.1126/science.1193225
Spiroindolones, a potent compound class for the treatment of malaria
Abstract
Recent reports of increased tolerance to artemisinin derivatives--the most recently adopted class of antimalarials--have prompted a need for new treatments. The spirotetrahydro-beta-carbolines, or spiroindolones, are potent drugs that kill the blood stages of Plasmodium falciparum and Plasmodium vivax clinical isolates at low nanomolar concentration. Spiroindolones rapidly inhibit protein synthesis in P. falciparum, an effect that is ablated in parasites bearing nonsynonymous mutations in the gene encoding the P-type cation-transporter ATPase4 (PfATP4). The optimized spiroindolone NITD609 shows pharmacokinetic properties compatible with once-daily oral dosing and has single-dose efficacy in a rodent malaria model.
Figures

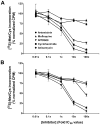
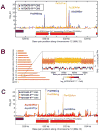

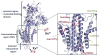
Comment in
-
Microbiology. Is the tide turning for new malaria medicines?Science. 2010 Sep 3;329(5996):1153-4. doi: 10.1126/science.1194923. Science. 2010. PMID: 20813940 No abstract available.
-
Antimalarial drugs: Speeding to a new lead.Nat Rev Drug Discov. 2010 Nov;9(11):842. doi: 10.1038/nrd3301. Nat Rev Drug Discov. 2010. PMID: 21031000 No abstract available.
References
-
- WHO. World Malaria Report. 2009. http://www.who.int/malaria/world_malaria_report_2009/en/
-
- Noedl H, et al. N Engl J Med. 2008;359:2619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical
Molecular Biology Databases

