Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I
- PMID: 20814424
- PMCID: PMC2948184
- DOI: 10.1038/embor.2010.130
Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I
Abstract
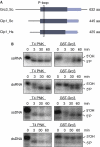
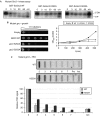
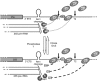
Transcription termination by RNA polymerase I in Saccharomyces cerevisiae is mediated by a 'torpedo' mechanism: co-transcriptional RNA cleavage by Rnt1 at the ribosomal DNA 3'-region generates a 5'-end that is recognized by the 5'-3' exonuclease Rat1; this degrades the downstream transcript and eventually causes termination. In this study, we identify Grc3 as a new factor involved in this process. We demonstrate that GRC3, an essential gene of previously unknown function, encodes a polynucleotide kinase that is required for efficient termination by RNA polymerase I. We propose that it controls the phosphorylation status of the downstream Rnt1 cleavage product and thereby regulates its accessibility to the torpedo Rat1.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- El-Moghazy AN et al. (2000) Functional analysis of six novel ORFs on the left arm of chromosome XII in Saccharomyces cerevisiae reveals two essential genes, one of which is under cell-cycle control. Yeast 16: 277–288 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

