Hydrophobic core formation and dehydration in protein folding studied by generalized-ensemble simulations
- PMID: 20816077
- PMCID: PMC2931739
- DOI: 10.1016/j.bpj.2010.06.045
Hydrophobic core formation and dehydration in protein folding studied by generalized-ensemble simulations
Abstract
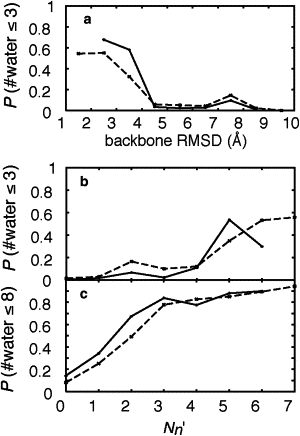
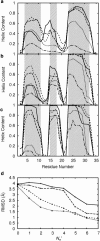
Despite its small size, chicken villin headpiece subdomain HP36 folds into the native structure with a stable hydrophobic core within several microseconds. How such a small protein keeps up its conformational stability and fast folding in solution is an important issue for understanding molecular mechanisms of protein folding. In this study, we performed multicanonical replica-exchange simulations of HP36 in explicit water, starting from a fully extended conformation. We observed at least five events of HP36 folding into nativelike conformations. The smallest backbone root mean-square deviation from the crystal structure was 1.1 A. In the nativelike conformations, the stably formed hydrophobic core was fully dehydrated. Statistical analyses of the simulation trajectories show the following sequential events in folding of HP36: 1), Helix 3 is formed at the earliest stage; 2), the backbone and the side chains near the loop between Helices 2 and 3 take nativelike conformations; and 3), the side-chain packing at the hydrophobic core and the dehydration of the core side chains take place simultaneously at the later stage of folding. This sequence suggests that the initial folding nucleus is not necessarily the same as the hydrophobic core, consistent with a recent experimental phi-value analysis.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Salt effects on hydrophobic-core formation in folding of a helical miniprotein studied by molecular dynamics simulations.Proteins. 2014 Jun;82(6):933-43. doi: 10.1002/prot.24467. Epub 2013 Dec 6. Proteins. 2014. PMID: 24214490
-
Contact pair dynamics during folding of two small proteins: chicken villin head piece and the Alzheimer protein beta-amyloid.J Chem Phys. 2004 Jan 15;120(3):1602-12. doi: 10.1063/1.1633253. J Chem Phys. 2004. PMID: 15268287
-
Reconciling the solution and X-ray structures of the villin headpiece helical subdomain: molecular dynamics simulations and double mutant cycles reveal a stabilizing cation-pi interaction.Biochemistry. 2007 Mar 27;46(12):3624-34. doi: 10.1021/bi061785+. Epub 2007 Mar 6. Biochemistry. 2007. PMID: 17338549 Free PMC article.
-
Characterizing a partially ordered miniprotein through folding molecular dynamics simulations: Comparison with the experimental data.Protein Sci. 2016 Mar;25(3):587-96. doi: 10.1002/pro.2850. Epub 2015 Dec 16. Protein Sci. 2016. PMID: 26609791 Free PMC article.
-
Effect of subdomain interactions on methyl group dynamics in the hydrophobic core of villin headpiece protein.Protein Sci. 2014 Feb;23(2):145-56. doi: 10.1002/pro.2398. Epub 2013 Dec 3. Protein Sci. 2014. PMID: 24243806 Free PMC article.
Cited by
-
Short disordered protein segment regulates cross-species transmission of a yeast prion.Nat Chem Biol. 2020 Jul;16(7):756-765. doi: 10.1038/s41589-020-0516-y. Epub 2020 Apr 13. Nat Chem Biol. 2020. PMID: 32284601
-
Decoding Structural Properties of a Partially Unfolded Protein Substrate: En Route to Chaperone Binding.PLoS Comput Biol. 2015 Sep 22;11(9):e1004496. doi: 10.1371/journal.pcbi.1004496. eCollection 2015. PLoS Comput Biol. 2015. PMID: 26394388 Free PMC article.
-
Solvent-Driven Dynamical Crossover in the Phenylalanine Side-Chain from the Hydrophobic Core of Amyloid Fibrils Detected by 2H NMR Relaxation.J Phys Chem B. 2017 Aug 3;121(30):7267-7275. doi: 10.1021/acs.jpcb.7b04726. Epub 2017 Jul 21. J Phys Chem B. 2017. PMID: 28699757 Free PMC article.
-
Protein structure predictions by enhanced conformational sampling methods.Biophys Physicobiol. 2019 Nov 29;16:344-366. doi: 10.2142/biophysico.16.0_344. eCollection 2019. Biophys Physicobiol. 2019. PMID: 31984190 Free PMC article. Review.
-
Flexible Proteins at the Origin of Life.Life (Basel). 2017 Jun 5;7(2):23. doi: 10.3390/life7020023. Life (Basel). 2017. PMID: 28587235 Free PMC article.
References
-
- Shen M., Davis F.P., Sali A. The optimal size of a globular protein domain: A simple sphere-packing model. Chem. Phys. Lett. 2005;405:224–228.
-
- Veretnik S., Bourne P.E., Shindyalov I.N. Toward consistent assignment of structural domains in proteins. J. Mol. Biol. 2004;339:647–678. - PubMed
-
- McKnight C.J., Doering D.S., Kim P.S. A thermostable 35-residue subdomain within villin headpiece. J. Mol. Biol. 1996;260:126–134. - PubMed
-
- Wang M., Tang Y., Raleigh D.P. Dynamic NMR line-shape analysis demonstrates that the villin headpiece subdomain folds on the microsecond time scale. J. Am. Chem. Soc. 2003;125:6032–6033. - PubMed
-
- Kubelka J., Eaton W.A., Hofrichter J. Experimental tests of villin subdomain folding simulations. J. Mol. Biol. 2003;329:625–630. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

