BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies
- PMID: 20817139
- PMCID: PMC2933337
- DOI: 10.1016/j.ajhg.2010.07.021
BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies
Abstract
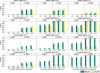
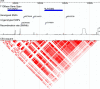
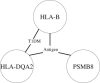
Gene-gene interactions have long been recognized to be fundamentally important for understanding genetic causes of complex disease traits. At present, identifying gene-gene interactions from genome-wide case-control studies is computationally and methodologically challenging. In this paper, we introduce a simple but powerful method, named "BOolean Operation-based Screening and Testing" (BOOST). For the discovery of unknown gene-gene interactions that underlie complex diseases, BOOST allows examination of all pairwise interactions in genome-wide case-control studies in a remarkably fast manner. We have carried out interaction analyses on seven data sets from the Wellcome Trust Case Control Consortium (WTCCC). Each analysis took less than 60 hr to completely evaluate all pairs of roughly 360,000 SNPs on a standard 3.0 GHz desktop with 4G memory running the Windows XP system. The interaction patterns identified from the type 1 diabetes data set display significant difference from those identified from the rheumatoid arthritis data set, although both data sets share a very similar hit region in the WTCCC report. BOOST has also identified some disease-associated interactions between genes in the major histocompatibility complex region in the type 1 diabetes data set. We believe that our method can serve as a computationally and statistically useful tool in the coming era of large-scale interaction mapping in genome-wide case-control studies.
2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bateson W., Mendel G. Cambridge University Press; Cambridge: 1909. Mendel's Principles of Heredity.
-
- Fisher R.A. The correlations between relatives on the supposition of mendelian inheritance. Philosophical Transactions of the Royal Society of Edinburgh. 1918;52:399–433.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

