Structures of the lipopolysaccharides from Rhizobium leguminosarum RBL5523 and its UDP-glucose dehydrogenase mutant (exo5)
- PMID: 20817634
- PMCID: PMC2998983
- DOI: 10.1093/glycob/cwq131
Structures of the lipopolysaccharides from Rhizobium leguminosarum RBL5523 and its UDP-glucose dehydrogenase mutant (exo5)
Abstract
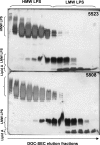
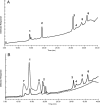
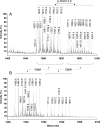
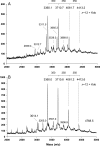
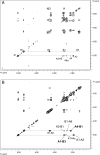
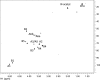
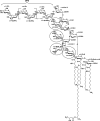
Rhizobial lipopolysaccharide (LPS) is required to establish an effective symbiosis with its host plant. An exo5 mutant of Rhizobium leguminosarum RBL5523, strain RBL5808, is defective in UDP-glucose (Glc) dehydrogenase that converts UDP-Glc to UDP-glucuronic acid (GlcA). This mutant is unable to synthesize either UDP-GlcA or UDP-galacturonic acid (GalA) and is unable to synthesize extracellular and capsular polysaccharides, lacks GalA in its LPS and is defective in symbiosis (Laus MC, Logman TJ, van Brussel AAN, Carlson RW, Azadi P, Gao MY, Kijne JW. 2004. Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol. 186:6617-6625). Here, we determined and compared the structures of the RBL5523 parent and RBL5808 mutant LPSs. The parent LPS core oligosaccharide (OS), as with other R. leguminosarum and Rhizobium etli strains, is a Gal(1)Man(1)GalA(3)Kdo(3) octasaccharide in, which each of the GalA residues is terminally linked. The core OS from the mutant lacks all three GalA residues. Also, the parent lipid A consists of a fatty acylated GlcNGlcNonate or GlcNGlcN disaccharide that has a GalA residue at the 4'-position, typical of other R. leguminosarum and R. etli lipids A. The mutant lipid A lacks the 4'-GalA residue, and the proximal glycosyl residue was only present as GlcNonate. In spite of these alterations to the lipid A and core OSs, the mutant was still able to synthesize an LPS containing a normal O-chain polysaccharide (OPS), but at reduced levels. The structure of the OPS of the mutant LPS was identical to that of the parent and consists of an O-acetylated →4)-α-d-Glcp-(1→3)-α-d-QuipNAc-(1→ repeating unit.
Figures




References
-
- Bhat UR, Forsberg LS, Carlson RW. Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide: unique non-phosphorylated lipid A containing 2-amino-2-deoxy-gluconate, galacturonate, and glucosamine. J Biol Chem. 1994;269:14402–14410. - PubMed
-
- Carlson RW, Forsberg LS, Kannenberg EL. Lipopolysaccharides in Rhizobium-legume symbioses. In: Quinn PJ, Wang X, editors. Subcellular Biochemistry. Endotoxins: Structure, Function and Recognition. vol. 53. Secaucus, NJ: Springer; 2010. pp. 339–386. - PubMed
-
- Carlson RW, Garci F, Noel D, Hollingsworth R. The structures of the lipopolysaccharide core components from Rhizobium leguminosarum biovar phaseoli CE3 and two of its symbiotic mutants, CE109 and CE309. Carbohydr Res. 1989;195:101–110. doi:10.1016/0008-6215(89)85092-X. - DOI - PubMed

